Distinct Expression Profiles of Three Melatonin Receptors during Early Development and Metamorphosis in the Flatfish Solea senegalensis
Abstract
:1. Introduction
2. Results
2.1. mt1 Expression
| mt1 | |||||||||||||||||||||||||||||||
| TWO-WAY ANOVA | KRUSKAL WALLIS | ||||||||||||||||||||||||||||||
| Developmental stage | Hour of sampling | Interaction | H = 60.4; p < 0.0001 | ||||||||||||||||||||||||||||
| F = 10.83; p < 0.001 | F = 23.6; p < 0.001 | F = 3.15; p < 0.01 | |||||||||||||||||||||||||||||
| BONFERRONI TEST | |||||||||||||||||||||||||||||||
| Developmental stage | UE | 0 dpf | 2 dpf | 4 dpf | 6 dpf | 9 dpf | 12 dpf | 15 dpf | 19 dpf | 21 dpf | |||||||||||||||||||||
| Hour of sampling | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ||||||||||||
| Comparison | a | abcd | ab | ab | abcd | abcd | cde | abcd | e | abcd | bcde | abcd | e | abcd | de | abc | abcd | ab | abc | ||||||||||||
| mt2 | |||||||||||||||||||||||||||||||
| TWO-WAY ANOVA | KRUSKAL WALLIS | ||||||||||||||||||||||||||||||
| Developmental stage | Hour of sampling | Interaction | H = 57.7; p < 0.0001 | ||||||||||||||||||||||||||||
| F = 10.67; p < 0.001 | F = 0.10; p = 0.756 | F = 2.29; p < 0.05 | |||||||||||||||||||||||||||||
| BONFERRONI TEST | |||||||||||||||||||||||||||||||
| Developmental stage | UE | 0 dpf | 2 dpf | 4 dpf | 6 dpf | 9 dpf | 12 dpf | 15 dpf | 19 dpf | 21 dpf | |||||||||||||||||||||
| Hour of sampling | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ML | MD | ||||||||||||
| Comparison | a | abc | a | abcdefg | fg | defg | cdefg | g | bcdefg | efg | abcdefg | abcdef | abcdef | abcde | abcde | abcd | abcdef | abcd | abcde | ||||||||||||
| mel1c | |||||||||||||||||||||||||||||||
| TWO-WAY ANOVA | |||||||||||||||||||||||||||||||
| Developmental stage | Hour of sampling | Interaction | |||||||||||||||||||||||||||||
| F = 6.54; p < 0.001 | F = 16.87; p < 0.001 | F = 0.44; p = 0.8729 | |||||||||||||||||||||||||||||
| NEWMAN-KEULS TEST | |||||||||||||||||||||||||||||||
| Developmental stage | UE | 0 dpf | 2 dpf | 4 dpf | 6 dpf | 9 dpf | 12 dpf | 15 dpf | 19 dpf | 21 dpf | |||||||||||||||||||||
| Comparison | a | N/D | bc | bc | b | bc | c | c | b | b | |||||||||||||||||||||
| Hour of sampling | ML | MD | |||||||||||||||||||||||||||||
| Comparison | a | b | |||||||||||||||||||||||||||||
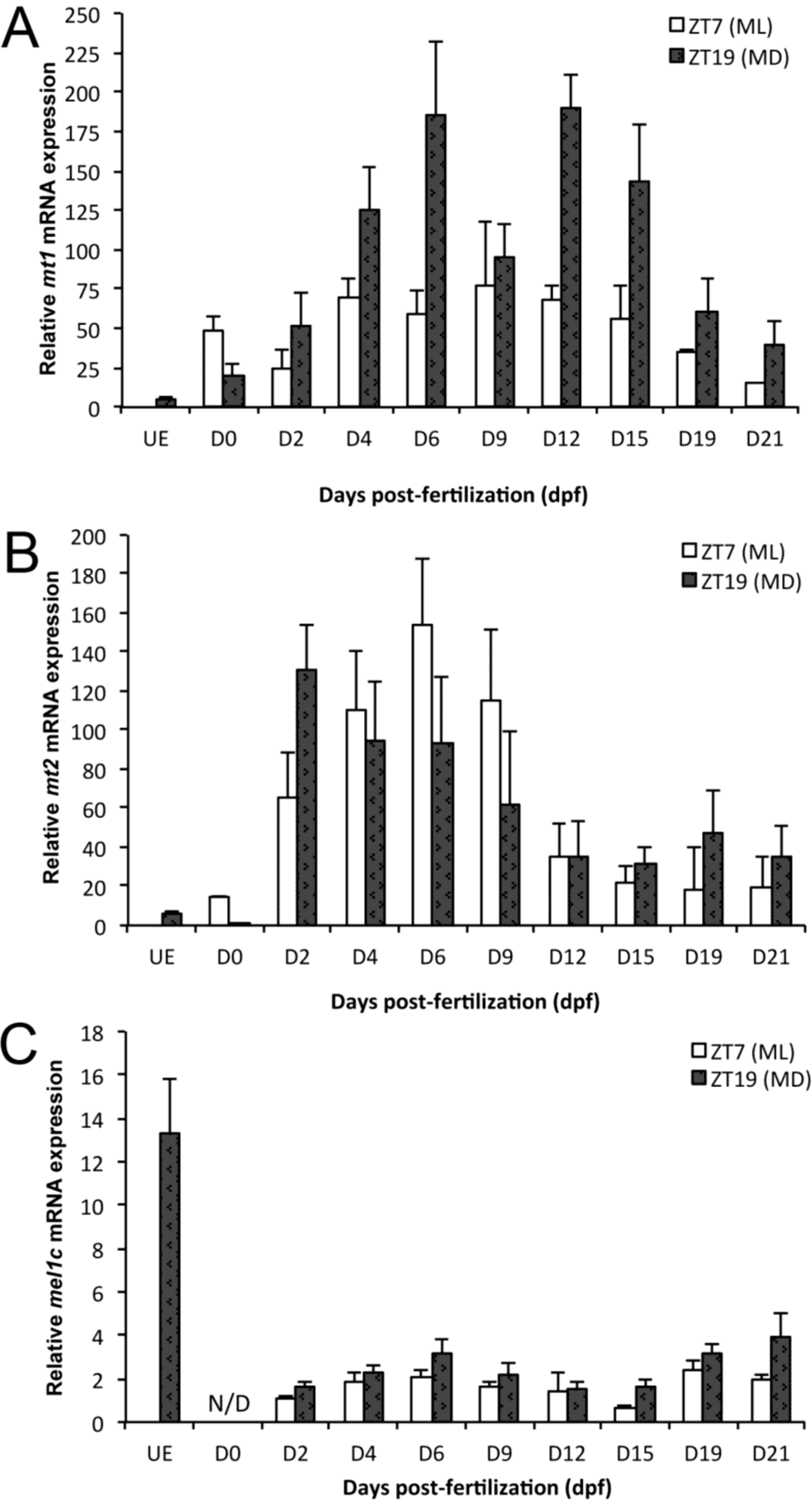
2.2. mt2 Expression
2.3. mel1c Expression
3. Discussion
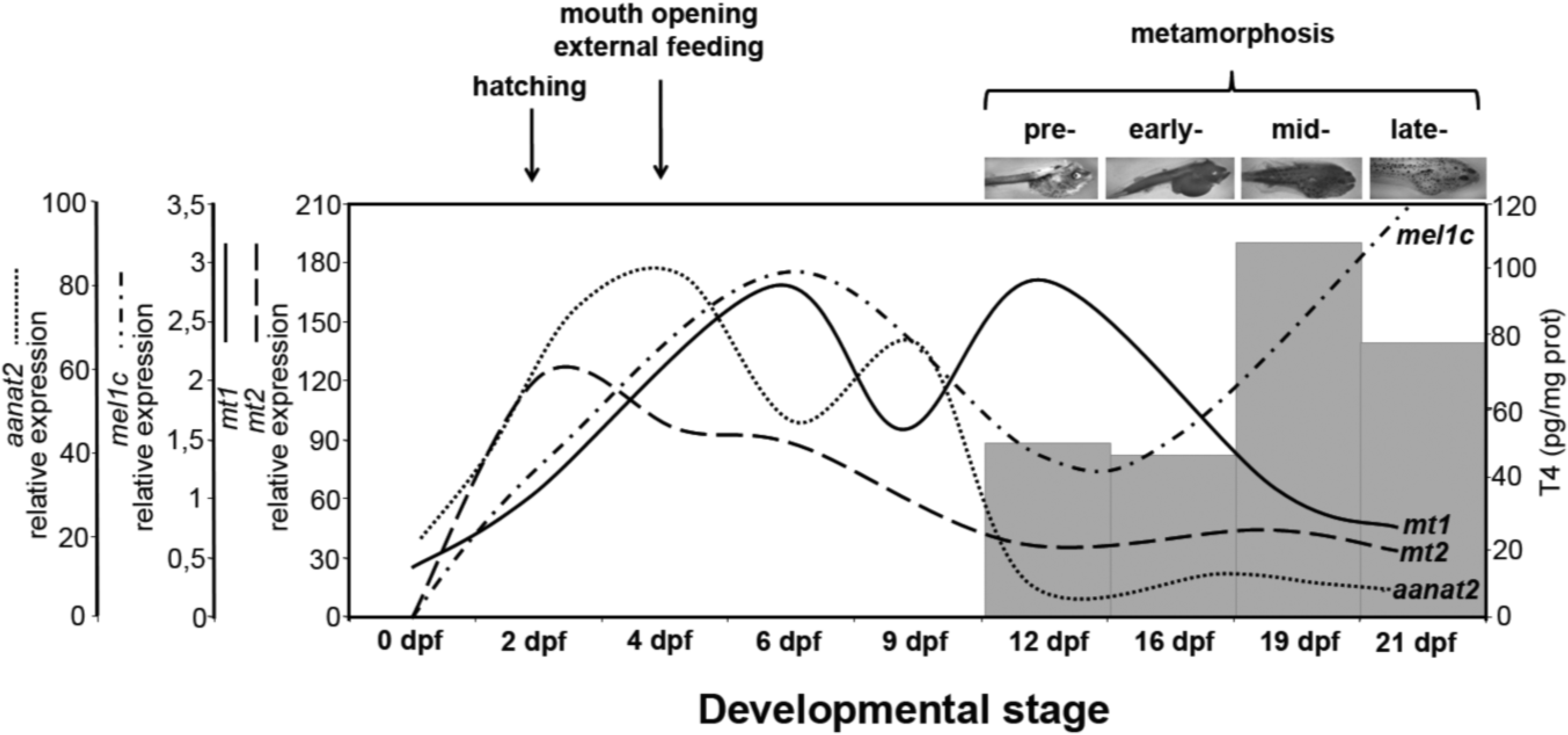
4. Materials and Methods
4.1. Animals and Sampling
4.2. Quantitative Real-Time PCR Analysis
| Primer Name | Sequence (5'-3') |
|---|---|
| ssmt1F | GCGGAAAGGAATAAATGAGGC |
| ssmt1R | GGAGTTGGTGCGTCACAGTG |
| ssmt2F | GCGTCAACGAAGAGCGAAAT |
| ssmt2R | GCCCGAAACTGGCCATAAAT |
| ssmel1cF | ACTTCAACAGCTGCCTCAACG |
| ssmel1cR | AGCAAACGTGGGATGCAAAG |
| ssβactinF | GGATCTGCATGCCAACACTG |
| ssβactinR | TCTGCATCCTGTCAGCAATG |
4.3. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reiter, R.J. The melatonin rhythm. Experientia 1993, 49, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Falcón, J.; Migaud, H.; Muñoz-Cueto, J.A.; Carrillo, M. Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 469–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reppert, S.M.; Weave, D.R.; Godson, C. Melatonin receptors step into the light: cloning and classification of subtypes. Trends Pharmacol. Sci. 1996, 17, 100–102. [Google Scholar] [CrossRef] [PubMed]
- Sauzet, S.; Besseau, L.; Herrera-Perez, P.; Covès, D.; Chatain, B.; Peyric, E.; Boeuf, G.; Muñoz-Cueto, J.A.; Falcón, J. Cloning and retinal expression of melatonin receptors in the European sea bass, Dicentrarchus labrax. Gen. Comp. Endocrinol. 2008, 157, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Pérez, P.; Rendón, M.C.; Besseau, L.; Sauzet, S.; Falcón, J.; Muñoz-Cueto, J.A. Melatonin receptors in the brain of the European sea bass: An in situ hybridization and autoradiographic study. J. Comp. Neurol. 2010, 518, 3495–3511. [Google Scholar] [CrossRef] [PubMed]
- Danilova, N.; Krupnik, V.E.; Sugden, D.; Zhdanova, I.V. Melatonin stimulates cell proliferation in zebrafish embryo and accelerates its development. FASEB J. 2004, 18, 751–753. [Google Scholar] [PubMed]
- Ziv, L.; Gothilf, Y. Circadian time-keeping during early stages of development. Proc. Natl. Acad. Sci. USA 2006, 103, 4146–4151. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.W.; Easter, S.S., Jr. Stereotyped pathway selection by growth cones of early epiphysial neurons in the embryonic zebrafish. Development 1991, 112, 723–746. [Google Scholar] [PubMed]
- Forsell, J.; Holmqvist, B.; Helvik, J.V.; Ekström, P. Role of the pineal organ in the photoregulated hatching of the Atlantic halibut. Int. J. Dev. Biol. 1997, 41, 591–595. [Google Scholar] [PubMed]
- Vuilleumier, R.; Besseau, L.; Boeuf, G.; Piparelli, A.; Gothilf, Y.; Gehring, W.G.; Klein, D.C.; Falcón, J. Starting the zebrafish pineal circadian clock with a single photic transition. Endocrinology 2006, 147, 2273–2279. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.L. Melatonin, diel rhythms, and metamorphosis in anuran amphibians. Gen. Comp. Endocrinol. 2002, 126, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Isorna, E.; Guijarro, A.I.; Delgado, M.J.; López-Patiño, M.A.; de Pedro, N.; Alonso-Gómez, A.L. Ontogeny of central melatonin receptors in tadpoles of the anuran Rana perezi: modulation of dopamine release. J. Comp. Physiol. A 2005, 191, 1099–1105. [Google Scholar] [CrossRef]
- Shi, Q.; Ando, H.; Coon, S.L.; Sato, S.; Ban, M.; Urano, A. Embryonic and post-embryonic expression of arylalkylamine N-acetyltransferase and melatonin receptor genes in the eye and brain of chum salmon (Oncorhynchus keta). Gen. Comp. Endocrinol. 2004, 136, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Bayarri, M.J.; Muñoz-Cueto, J.A.; López-Olmeda, J.F.; Vera, L.M.; Rol de Lama, M.A.; Madrid, J.A.; Sánchez-Vázquez, F.J. Daily locomotor activity and melatonin rhythms in Senegal sole (Solea senegalensis). Physiol. Behav. 2004, 81, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Vera, L.M.; Oliveira, C.; López-Olmeda, J.F.; Ramos, J.; Mañanos, E.; Madrid, J.A.; Sánchez-Vázquez, F.J. Seasonal and daily plasma melatonin rhythms and reproduction in Senegal sole kept under natural photoperiod and natural or controlled water temperature. J. Pineal Res. 2007, 43, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Isorna, E.; El M’Rabet, A.; Confente, F.; Falcón, J.; Muñoz-Cueto, J.A. Cloning and expression of arylalkylamine N-acetyltranferase-2 during early development and metamorphosis in the sole Solea senegalensis. Gen. Comp. Endocrinol. 2009, 161, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Isorna, E.; Aliaga-Guerrero, M.; El M’Rabet, A.; Servili, A.; Falcón, J.; Muñoz-Cueto, J.A. Identification of two arylalkylamine N-acetyltranferase 1 genes with different developmental expression profiles in the flatfish Solea senegalensis. J. Pineal Res. 2011, 51, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Confente, F.; Rendón, M.C.; Besseau, L.; Falcón, J.; Muñoz-Cueto, J.A. Melatonin receptors in a pleuronectiform species, Solea senegalensis: Cloning, tissue expression, day-night and seasonal variations. Gen. Comp. Endocrinol. 2010, 167, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Obłap, R.; Olszanska, B. Expression of melatonin receptor transcripts (mel-1a, mel-1b and mel-1c) in Japanese quail oocytes and eggs. Zygote 2001, 9, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Olszanska, B.; Majewski, P.; Lewczuk, B.; Stepinska, U. Melatonin and its synthesizing enzymes (arylalkylamine N-acetyltransferase-like and hydroxyindole-O-methyltransferase) in avian eggs and early embryos. J. Pineal Res. 2007, 42, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Dinis, M.T.; Soares, F.; Cabrita, E.; Pousão-Ferreira, P.; Sánchez-Vázquez, F.J. Lunar and daily spawning rhythms of Senegal sole Solea senegalensis. J. Fish. Biol. 2009, 75, 61–74. [Google Scholar] [CrossRef] [PubMed]
- El M’Rabet, A.; Confente, F.; Ouarour, A.; Muñoz-Cueto, J.A. Ontogenia del órgano pineal del lenguado, Solea senegalensis. In Avances en Endocrinología Comparada; Muñoz-Cueto, J.A., Mancera, J.M., Martínez, G., Eds.; Cádiz: Servicio de Publicaciones de la Universidad de Cádiz, Cádiz, Spain, 2008; Volume 4, pp. 167–170. [Google Scholar]
- Blanco-Vives, B.; Aliaga-Guerrero, M.; Cañavate, J.P.; Muñoz-Cueto, J.A.; Sánchez-Vázquez, F.J. Does lighting manipulation during incubation affect hatching rhythms and early development of sole? Chronobiol. Int. 2011, 28, 300–306. [Google Scholar]
- Cañavate, J.P.; Zerolo, R.; Fernández-Díaz, C. Feeding and development of Senegal sole (Solea senegalensis) larvae reared in different photoperiods. Aquaculture 2006, 258, 368–377. [Google Scholar] [CrossRef]
- Isorna, E.; Obregon, M.J.; Calvo, R.M.; Vázquez, R.; Pendón, C.; Falcón, J.; Muñoz-Cueto, J.A. Iodothyronine deiodinases and thyroid hormone receptors regulation during flatfish (Solea senegalensis) metamorphosis. J. Exp. Zool. B 2009, 312, 231–246. [Google Scholar] [CrossRef]
- Power, D.M.; Llewellyn, L.; Faustino, M.; Nowell, M.A.; Bjornsson, B.T.; Einarsdottir, I.E.; Canario, A.V.M.; Sweeney, G.E. Thyroid hormones in growth and development of fish. Comp. Biochem. Physiol. C 2001, 130, 447–459. [Google Scholar]
- Kulczykowska, E.; Sokolowska, E.; Takvam, B.; Stefansson, S.; Ebbesson, L. Influence of exogenous thyroxine on plasma melatonin in juvenile Atlantic salmon (Salmo salar). Comp. Biochem. Physiol. B 2004, 137, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Ebisawa, T.; Karnes, S.; Lerner, M.R.; Reppert, S.M. Expression cloning of a high-affinity melatonin receptor from Xenopus dermal melanophores. Proc. Natl. Acad. Sci. USA 1994, 91, 6133–6137. [Google Scholar] [CrossRef]
- Vanecek, J. Cellular mechanisms of melatonin action. Physiol. Rev. 1998, 78, 687–721. [Google Scholar] [PubMed]
- Ikegami, T.; Azuma, K.; Nakamura, M.; Suzuki, N.; Hattori, A.; Ando, H. Diurnal expressions of four subtypes of melatonin receptor genes in the optic tectum and retina of goldfish. Comp. Biochem. Physiol. A 2009, 152, 219–224. [Google Scholar] [CrossRef]
- Aspengren, S.; Skold, H.N.; Quiroga, G.; Martensson, L.; Wallin, M. Noradrenaline- and melatonin-mediated regulation of pigment aggregation in fish melanophores. Pigment. Cell. Res. 2003, 16, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Dinis, M.T.; Ribeiro, L.; Soares, F.; Sarasquete, C. A review on the cultivation potential of Solea senegalensis in Spain and in Portugal. Aquaculture 1999, 176, 27–38. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan-Chow-Wing, O.; Confente, F.; Herrera-Pérez, P.; Isorna, E.; Chereguini, O.; Rendón, M.D.C.; Falcón, J.; Muñoz-Cueto, J.A. Distinct Expression Profiles of Three Melatonin Receptors during Early Development and Metamorphosis in the Flatfish Solea senegalensis. Int. J. Mol. Sci. 2014, 15, 20789-20799. https://doi.org/10.3390/ijms151120789
Lan-Chow-Wing O, Confente F, Herrera-Pérez P, Isorna E, Chereguini O, Rendón MDC, Falcón J, Muñoz-Cueto JA. Distinct Expression Profiles of Three Melatonin Receptors during Early Development and Metamorphosis in the Flatfish Solea senegalensis. International Journal of Molecular Sciences. 2014; 15(11):20789-20799. https://doi.org/10.3390/ijms151120789
Chicago/Turabian StyleLan-Chow-Wing, Olivier, Francesca Confente, Patricia Herrera-Pérez, Esther Isorna, Olvido Chereguini, Maria Del Carmen Rendón, Jack Falcón, and José A. Muñoz-Cueto. 2014. "Distinct Expression Profiles of Three Melatonin Receptors during Early Development and Metamorphosis in the Flatfish Solea senegalensis" International Journal of Molecular Sciences 15, no. 11: 20789-20799. https://doi.org/10.3390/ijms151120789





