Atrazine Affects Phosphoprotein and Protein Expression in MCF-10A Human Breast Epithelial Cells
Abstract
:1. Introduction
2. Results and Discussion
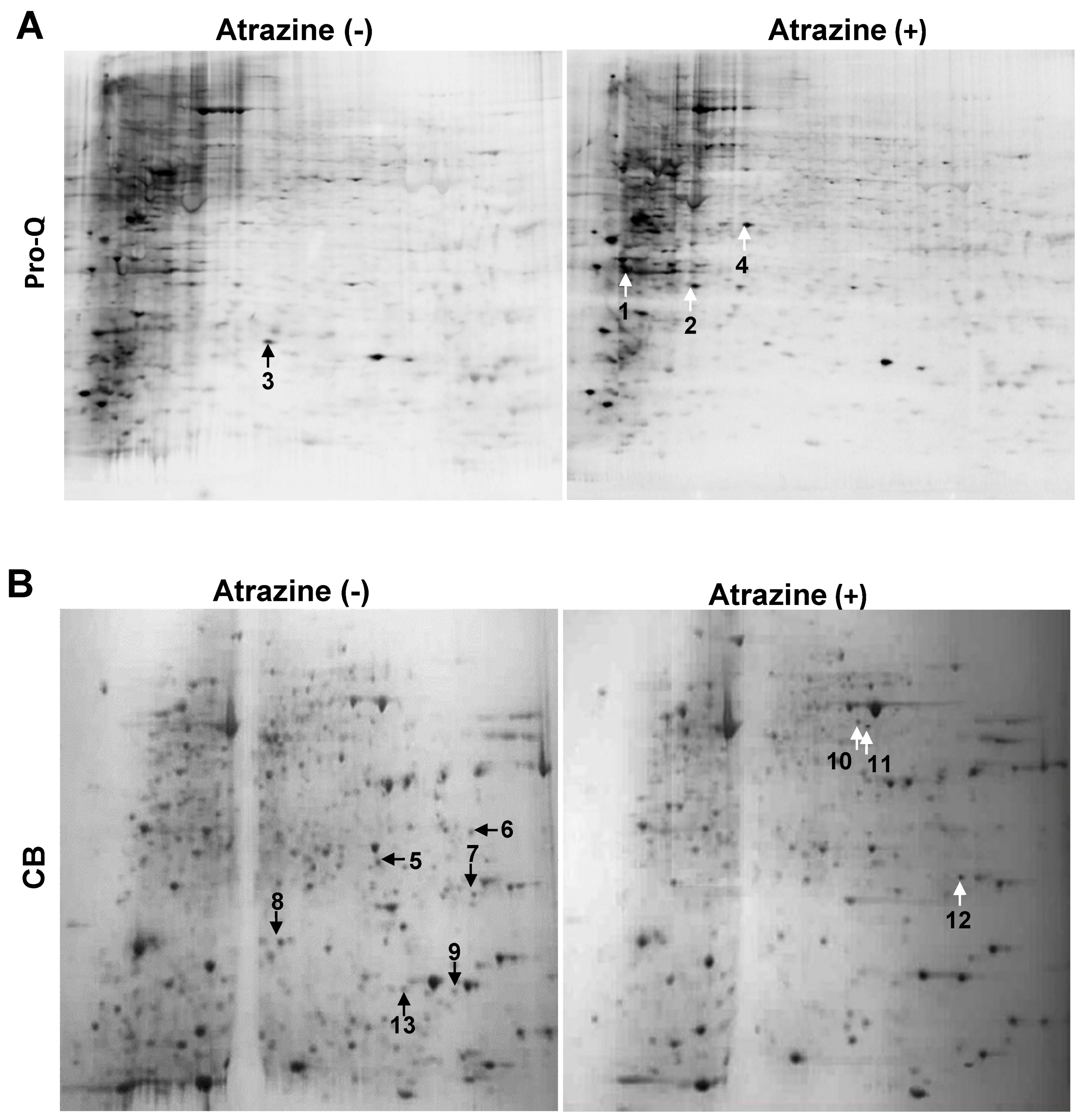
2.1. 2-DE Analysis
2.2. Protein Identification
| Spot # | Putative Protein Name | Name of Gene | Score | MW (kDa) | PI | Ion Score | Peptides |
|---|---|---|---|---|---|---|---|
| 1 | Acidic leucine-rich nuclear phosphoprotein 32 family, member A (gi: 5453880) | ANP32A | 129 | 29 | 3.99 | 29 | K.LLPQLTYLDGYDR.E |
| 85 | K.SLDLFNCEVTNLNDYR.E | ||||||
| 2 | Heat shock protein beta-1 (gi: 4504517) | HSPβ1 | 160 | 23 | 5.98 | 20 | R.GPSWDPER.D |
| 55 | R.LFDQAFGLPR.L | ||||||
| 3 | Nucloside Diphosphate Kinase A isoform b (gi: 4557797) | NME1 | 602 | 35 | 5.83 | 63 | R.GDFCIQVGR.N |
| 59 | K.DRPFFAGLVK.Y | ||||||
| 83 | R.TFIAIKPDGVQR.G | ||||||
| 94 | R.VMLGETNPADSKPGTIR.G | ||||||
| 4 | Pyruvate kinase, various isoforms (gi: 35505) | PDK | 425 | 58 | 7.58 | 63 | R.LDIDSPPITAR.N |
| 50 | R.LNFSHGTHEYHAETIK.N | ||||||
| 77 | K.IYVDDGLISLQVK.Q | ||||||
| 46 | R.RFDEILEASDGIMVAR.G | ||||||
| 5 | Tropomyosin alpha 3 chain isoform 2 (gi: 24119203) | TPM3 | 543 | 32 | 4.75 | 39 | K.LVIIEGDLER.T |
| 80 | R.IQLVEEELDR.A | ||||||
| 80 | R.KLVIIEGDLER.T | ||||||
| 29 | R.EQAEAEVASLNR.R | ||||||
| 6 | Guanine nucleotide-binding protein subunit beta-2-like 1 (gi: 5174447) | GNB2L1 | 646 | 35 | 7.6 | 67 | R.VWQVTIGTR.R |
| 77 | R.DETNYGIPQR.R | ||||||
| 133 | K.YTVQDESHSEWVSCVR.F | ||||||
| 82 | K.HLYTLDGGDIINALCFSPNR.Y | ||||||
| 7 | Transgelin-2 (gi: 4507357) | TAGLN2 | 232 | 23 | 8.4 | 42 | K.NVIGLQMGTNR.G |
| 76 | R.TLMNLGGLAVAR.D | ||||||
| 40 | K.QMEQISQFLQAAER.Y | ||||||
| 8 | Stathmin 1 (gi: 5031851) | STMN1 | 166 | 17 | 5.76 | 102 | R.ASGQAFELILSPR.S |
| 30 | R.SKESVPEFPLSPPK.K | ||||||
| 9 | Profilin-1 (gi: 4826898) | PFN1 | 177 | 15 | 8.4 | 41 | K.CYEMASHLR.R |
| 82 | R.DSLLQDGEFSMDLR.T | ||||||
| 10 | Heat shock protein 70 kDa protein 4 (gi: 62087882) | HSPA4 | 124 | 71 | 5.44 | 80 | K.DAGTIAGLNVLR.I |
| 11 | 219 | 59 | R.FEELNADLFR.G | ||||
| 78 | K.STAGDTHLGGEDFDNR.M | ||||||
| 103 | K.TVTNAVVTVPAYFNDSQR.Q | ||||||
| 12 | Peroxiredoxin-1 (gi: 4505591) | PRDX1 | 276 | 22 | 8.3 | 33 | K.ADEGISFR.G |
| 57 | K.IGHPAPNFK.A | ||||||
| 81 | R.QITVNDLPVGR.S | ||||||
| 28 | K.HGEVCPAGWKPGSDTIKPDVQK.S | ||||||
| 13 | 60S acidic ribosomal P12 (gi: 4506671) | RPS12 | 397 | 12 | 4.42 | 86 | K.ILDSVGIEADDDRLNK.V |
| 103 | R.YVASYLLAALGGNSSPSAK.D | ||||||
| 123 | K.LASVPAGGAVAVSAAPGSAAPAAGSAPAAAEEK.K |
| Gene name | Cellular localization | Category | Function | Expression (% control) | p value |
|---|---|---|---|---|---|
| NME1 | Cytoplasm; Nucleus | Metabolism | Major role in the synthesis of nucleoside triphosphates other than ATP | 32 | 0.031 |
| PDK | Mitochondrion | Plays a key role in glycolysis | 342 | 0.024 | |
| STMN1 | Cytoplasm; Cytoskeleton | Signalization | Involved in the regulation of microtubule (MT) | 27 | 0.047 |
| GNB2L1 | Cell/Peripheral membrane; Cytoplasm | Involved in the recruitment, assembly and/or regulation of a variety of signaling molecules | 23 | 0.035 | |
| RPS12 | Ribosome | Translation | Plays an important role in the elongation step of protein synthesis | 30 | 0.042 |
| ANP32A | Nucleus; Cytoplasm; ER | Transcription | Implicated in some cellular processes, including proliferation, differentiation, and apoptosis | 470 | 0.027 |
| PFN1 | Cytoplasm; Cytoskeleton | Structure | Binds to actin and affects the structure of the cytoskeleton | 21 | 0.033 |
| TAGLN2 | Nuclear/plasma membrane | Binds to actin, calmodulin, troponin C and tropomyosin | 19 | 0.014 | |
| TPM3 | Cytoplasm; Cytoskeleton | Binds to actin filaments in muscle and non-muscle cells | 33 | 0.039 | |
| PRDX1 | Cytoplasm | Catabolism | Involved in redox regulation of the cell | 260 | 0.040 |
| HSPβ1 | Cytoplasm | Chaperone | ATP-independent chaperone in protein folding | 327 | 0.012 |
| HSPA4 | Cytoplasm | HSP 70 kDa is a chaperone | 350 | 0.022 |
2.3. Classification of Identified Proteins
| Primer Name | Forward | Reverse |
|---|---|---|
| Transgelin-2 | 5'-TGGCTTTGGGCAGCTTGGCA-3' | 5'-TCCCTCCTGCAGCTGGCTCT-3' |
| Profilin-1 | 5'-CAAGAGCACCGGTGGGGCC-3' | 5'-GAACGCCGAAGGTGGGAGGC-3' |
| NDP kinase A | 5'-ACCGTCCATTCTTTGCCGGCC-3' | 5'-CGAGCATGACTCGGCCCGTC-3' |
| Pyruvate kinase | 5'-GCAGCGCCACTAAGCCGTGA-3' | 5'-AGCGGCCAGTTGTGGTCAGC-3' |
| ANP32A | 5'-TGCGGAACAGGACGCCCTCT-3' | 5'-TGCGATTGAGGTGAGGCCTACGT-3' |
| HSPβ1 | 5'-GTCGCAGTGGTTAGGCGGCA-3' | 5'-GACATCCAGGGACACGCGCC-3' |
| NAPDH (CK) | 5'-GCCTTCCGTGTCCCCACTGC-3' | 5'-CCTCCGACGCCTGCTTCACC-3' |
2.4. Atrazine Effects on the Phosphoproteome and Proteome of MCF-10A Human Cells
2.5. qPCR Verification of Transcriptional Regulation of the Identified Proteins
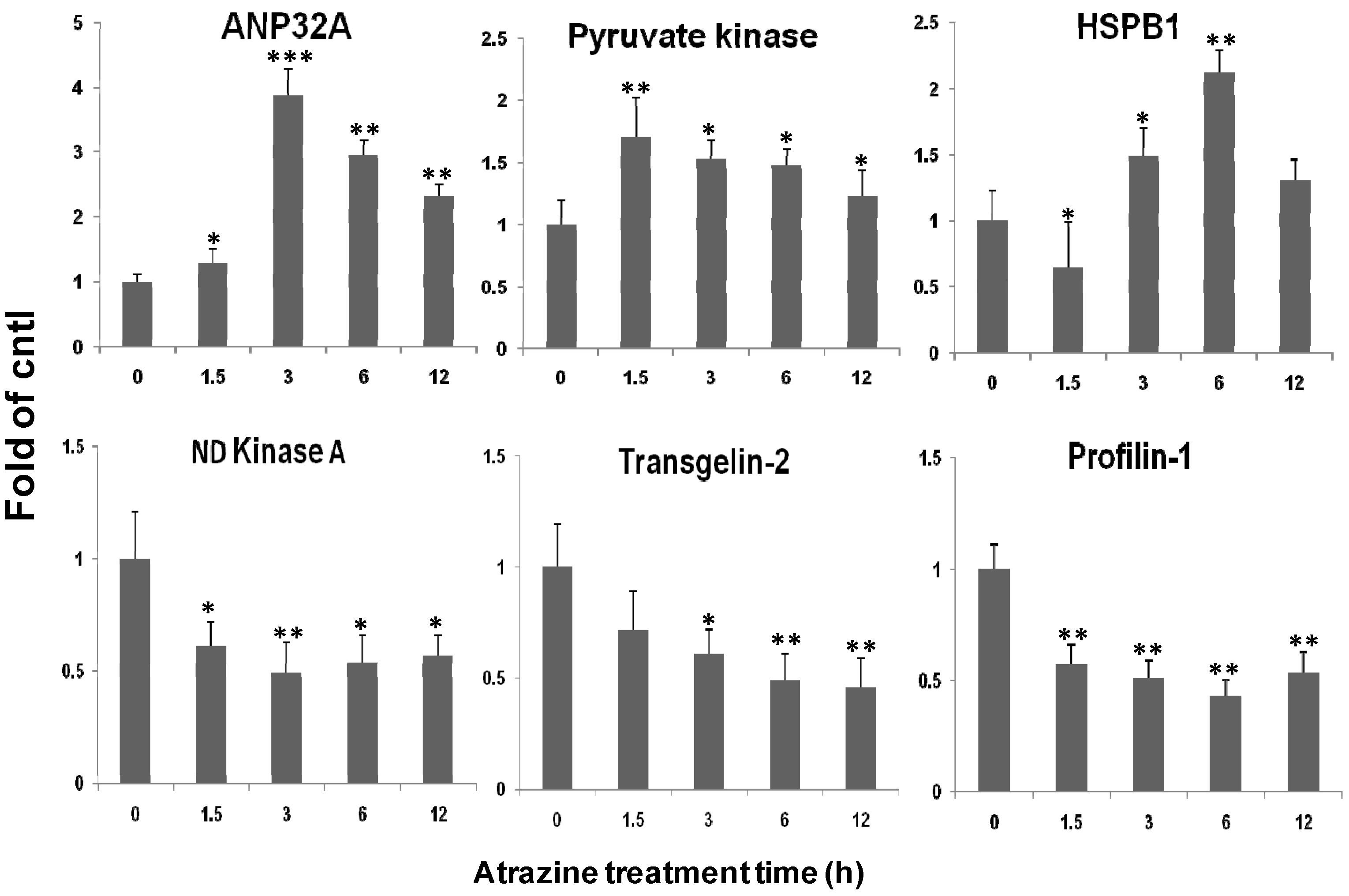
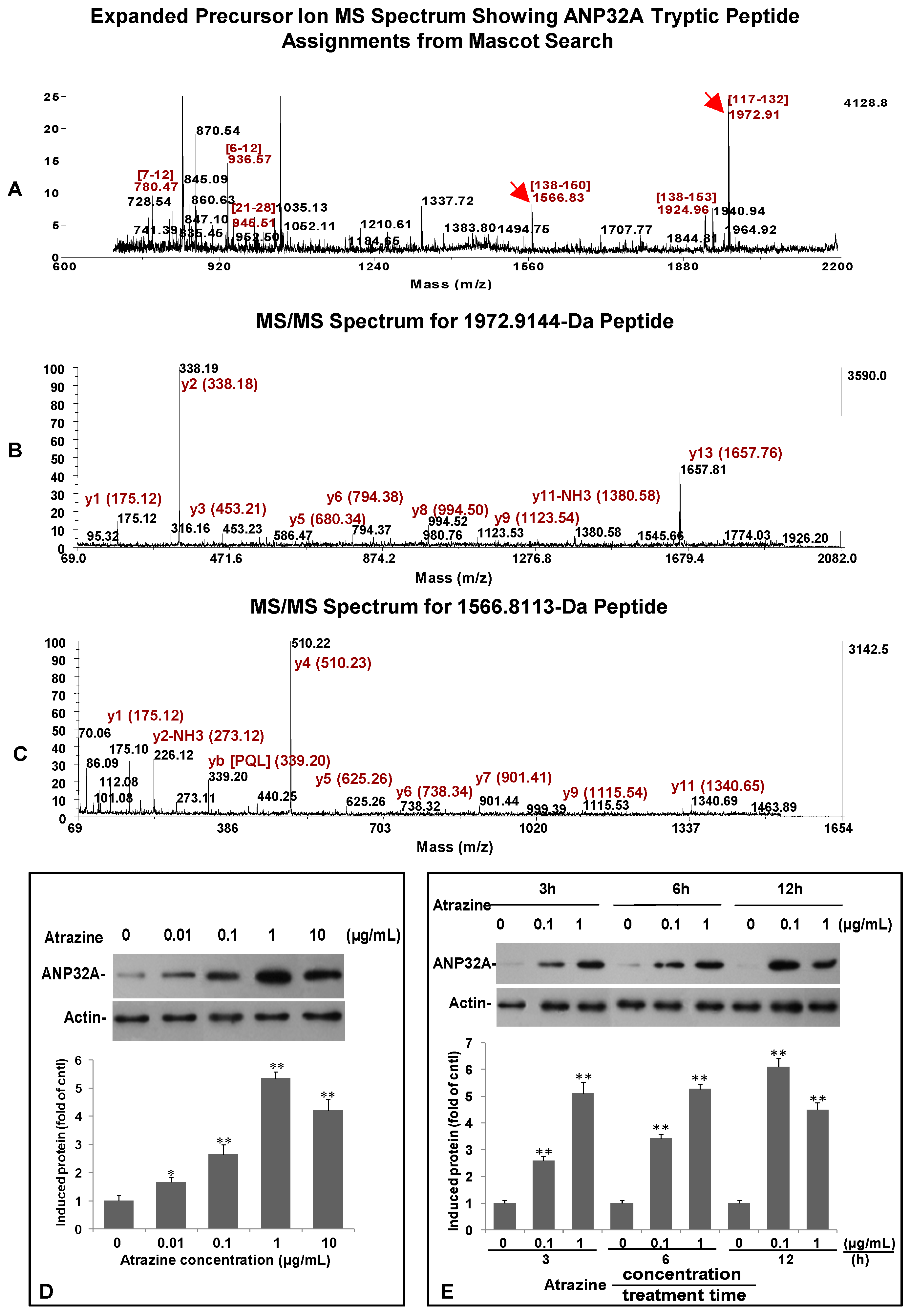
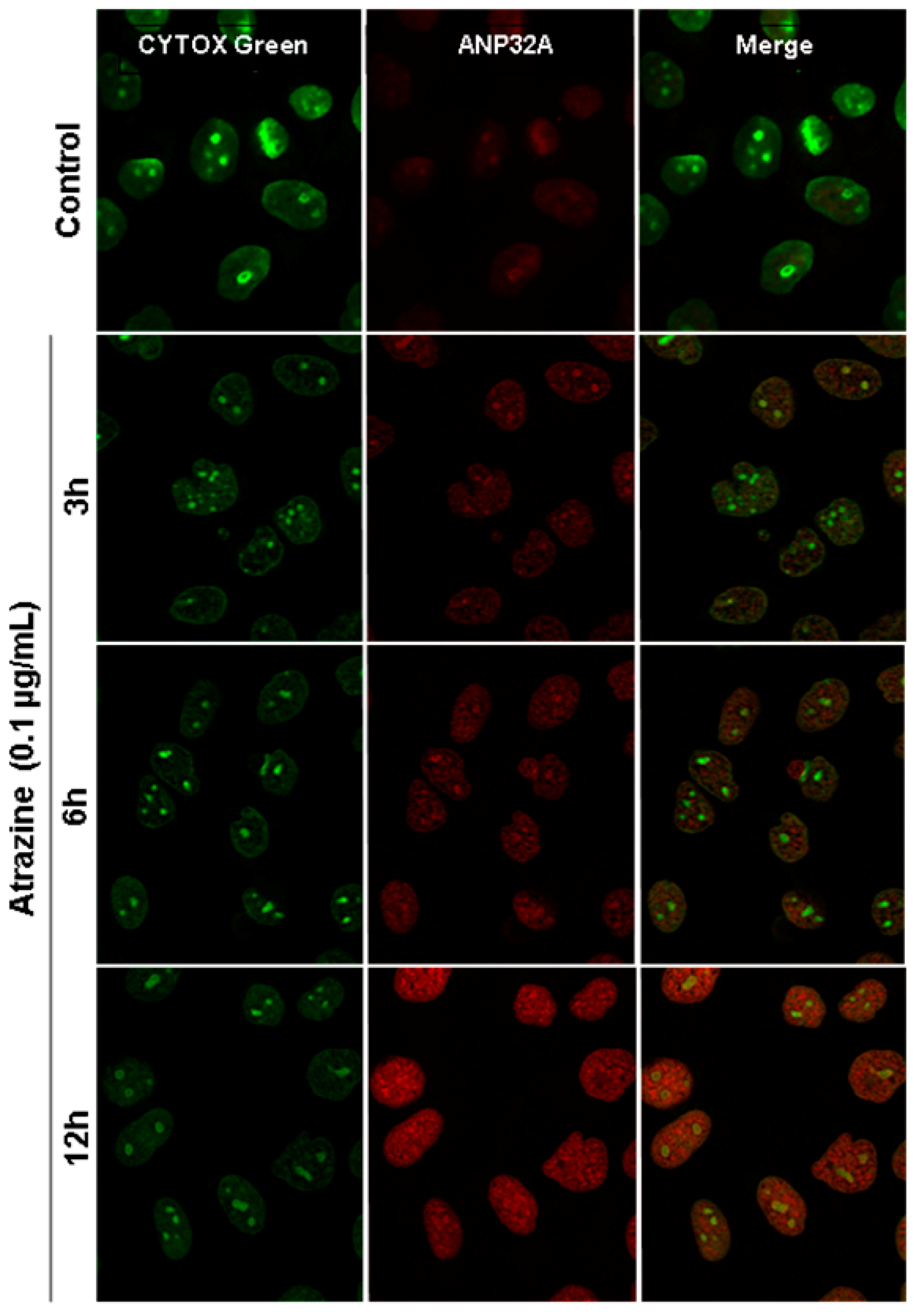
2.6. Verification of ANP32A Expression by Western blotting and Immunofluorescence Cell Staining
3. Experimental Section
3.1. Atrazine Preparation
3.2. Cell Culture
3.3. Identification of Differentially Expressed Proteins
3.4. In-Gel Protein Digestion
3.5. Trypsin Digest Processing and Sample Target Preparation
3.6. MALDI TOF/TOF MS and MS/MS Analysis
3.7. MALDI TOF/TOF MS Data Analysis
3.8. Quantitative Real-Time PCR (qPCR) Characterization
3.9. Western Blot Verification
3.10. Immunofluorescence Staining
3.11. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Short, P.; Colborn, T. Pesticide use in the U.S. and policy implications: A focus on herbicides. Toxicol. Ind. Health 1999, 15, 240–275. [Google Scholar] [CrossRef] [PubMed]
- Kiely, T.; Donaldson, D.; Grube, A. Pesticides Industry Sales and Usage: 2000 and 2001 Market Estimates; U.S. Environmental Protection Agency: Washington, DC, USA, 2004. [Google Scholar]
- Fenelon, J.; Moore, R. Transport of agrochemicals to ground and surface waters in a small central Indiana watershed. J. Environ. Qual. 1998, 27, 884–894. [Google Scholar] [CrossRef]
- Kolpin, D.; Barbash, J.; Gilliom, R. Occurrence of pesticides in shallow groundwater of the United States: Initial results from the National Water-Quality Assessment Program. Environ. Sci. Technol. 1998, 32, 558–566. [Google Scholar] [CrossRef]
- Lode, O.; Eklo, O.; Holen, B.; Svensen, A.; Johnsen, A. Pesticides in precipitation in Norway. Sci. Total Environ. 1995, 160, 421–431. [Google Scholar] [CrossRef]
- Miller, S.; Sweet, C.; Depinto, J.; Hornbuckle, K. Atrazine and nutrients in precipitation: Results from the Lake Michigan mass balance study. Environ. Sci. Technol. 2000, 34, 55–61. [Google Scholar] [CrossRef]
- Müller, S.; Berg, M.; Ulrich, M.; Schwarzenbach, R.P. Atrazine and its primary metabolites in Swiss lakes: Input characteristics and long-term behavior in the water column. Environ. Sci. Technol. 1997, 31, 2104–2113. [Google Scholar] [CrossRef]
- Solomon, K.; Giesy, J.; LaPoint, T.; Giddings, J.; Richards, R. Ecological risk assessment of atrazine in North American surface waters. Environ. Toxicol. Chem. 2013, 32, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Thurman, E.; Cromwell, A. Atmospheric transport, deposition, and fate of triazine herbicides and their metabolites in pristine areas at Isle Royale National Park. Environ. Sci. Technol. 2000, 34, 3079–3085. [Google Scholar] [CrossRef]
- Goldman, L.R. Atrazine, simazine, and cyanizine. In Notice of Initiation of Special Review, Fed Reg 59; U.S. Environmental Protection Agency: Washington, DC, USA, 1994; pp. 60412–60443. [Google Scholar]
- Selim, H. Retention and runoff losses of atrazine and metribuzin in soil. J. Environ. Qual. 2003, 32, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Gfrerer, M.; Martens, D.; Gawik, B.; Wenzi, T.; Zhang, A.; Quan, X.; Cheng, S.; Chen, J.; Platzer, B.; Lankmayr, E.; et al. Triazines in the aquatic systems of the eastern Chinese Rivers Liao-He and Yangtse. Chemosphere 2002, 47, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Dong, X.; Xie, H.; Wang, J.; Wang, J.; Su, J.; Yu, C. DNA damage and effects on glutathione-S transferase activity induced by atrazine exposure in zebrafish (Danio rerio). Environ. Toxicol. 2011, 26, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.L.; Stoker, T.E.; Tyrey, L.; Goldman, J.M.; McElroy, W.K. Atrazine disrupts the hypothalamic control of pituitary ovarian function. Toxicol. Sci. 2000, 53, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Coban, A.; Filipov, N.M. Dopaminergic toxicity associated with oral exposure to the herbicide atrazine in juvenile male C57BL/6 mice. J. Neurochem. 2007, 100, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, V.M.; Thiruchelvam, M.; Cory-Slechta, D.A. Sustained exposure to the widely used herbicide atrazine: Altered function and loss of neurons in brain monoamine systems. Environ. Health Perspect. 2005, 113, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Rayner, J.L.; Wood, C.; Fenton, S.E. Exposure parameters necessary for delayed puberty and mammary gland development in Long-Evans rats exposed in utero to atrazine. Toxicol. Appl. Pharmacol. 2004, 195, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Kniewald, J.; Jakominic, M.; Tomljenovic, A.; Simic, B.; Romac, P.; Vranesic, D.; Kniewald, Z. Disorders of male rat reproductive tract under the influence of atrazine. J. Appl. Toxicol. 2000, 20, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Hayes, T.; Haston, K.; Tsui, M.; Hoang, A.; Haeffele, C.; Vonk, A. Atrazine-induced hermaphroditism at 0.1 ppb in American leopard frogs (Rana pipiens): Laboratory and field evidence. Environ. Health Perspect. 2003, 111, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Hayes, T.B.; Stuart, A.A.; Mendoza, M.; Collins, A.; Noriega, N.; Vonk, A.; Johnston, G.; Liu, R.; Kpodzo, D. Characterization of atrazine-induced gonadal malformations in African clawed frogs (Xenopus laevis) and comparisons with effects of an androgen antagonist (cyproterone acetate) and exogenous estrogen (17 betaestradiol): Support for the demasculinization/feminization hypothesis. Environ. Health Perspect. 2006, 114, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Spano, L.; Tyler, C.R.; van Aerle, R.; Devos, P.; Mandiki, S.N.; Silvestre, F.; Thome, J.P.; Kestemont, P. Effects of atrazine on sex steroid dynamics, plasma vitellogenin concentration and gonad development in adult goldfish (Carassius auratus). Aquat. Toxicol. 2004, 66, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.M.; Brundage, K.M.; Barnett, J.B. Developmental immunotoxicity of atrazine in rodents. Basic Clin. Pharmacol. Toxicol. 2008, 102, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Filipov, N.M.; Pinchuk, L.M.; Boyd, B.L.; Crittenden, P.L. Immunotoxic effects of short-term atrazine exposure in young male C57BL/6 mice. Toxicol. Sci. 2005, 86, 324–332. [Google Scholar] [CrossRef]
- Betancourt, M.; Resendiz, A.; Fierro, E.C. Effect of two insecticides and two herbicides on the porcine sperm motility patterns using computer-assisted semen analysis (CASA) in vitro. Reprod. Toxicol. 2006, 22, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Tavera-Mendoza, L.; Ruby, S.; Brousseau, P.; Fournier, M.; Cyr, D.; Marcogliese, D. Response of the amphibian tadpole (Xenopus laevis) to atrazine during sexual differentiation of the testis. Environ. Toxicol. Chem. 2002, 21, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Donna, A.; Crosignani, P.; Robutti, F.; Betta, P.G.; Bocca, R.; Mariani, N.; Ferrario, F.; Fissi, R.; Berrino, F. Triazine herbicides and ovarian epithelial neoplasms. Scand. J. Work Environ. Health 1989, 15, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kettles, M.A.; Broning, S.R.; Prince, T.S.; Horstman, S.W. Triazine herbicide exposure and breast cancer incidence: An ecological study of Kentucky counties. Environ Health Perspect. 1997, 105, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Pintér, A.; Török, G.; Börzsönyi, M.; Surján, A.; Csík, M.; Kelecsényi, Z.; Kocsis, Z. Long-term carcinogenicity bioassay of the herbicide atrazine in F344 rats. Neoplasma 1990, 37, 533–544. [Google Scholar] [PubMed]
- Wetzel, L.T.; Luempert, L.G.; Breckenridge, C.B.; Tisdel, M.O.; Stevens, J.T.; Thakur, A.K.; Extrom, P.J.; Eldridge, J.C. Chronic effects of atrazine on estrus and mammary tumor formation in female Sprague-Dawley and Fischer 344 rats. J. Toxicol. Environ. Health 1994, 43, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Brawer, J.R.; Sonnenschein, C. Cytopathological effects of estradiol on the arcuate nucleus of the female rat. A possible mechanism for pituitary tumorigenesis. Am. J. Anat. 1975, 144, 57–88. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer (IARC), 1999. Summaries & Evaluation: ATRAZINE, 1999; 73, 59–67.
- Environmental Protection Agency. 2003; Interim registration eligibility decision for atrazine. Case No. 0062. EPA, Washington. [Google Scholar]
- Schmitz-Spanke, S.; Rettenmeier, A.W. Protein expression profiling in chemical carcinogenesis: A proteomic-based approach. Proteomics 2011, 11, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Lasserre, J.P.; Fack, F.; Revets, D.; Planchon, S.; Renaut, J.; Hoffmann, L.; Gutleb, A.C.; Muller, C.P.; Bohn, T. Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J. Proteome Res. 2009, 8, 5485–5496. [Google Scholar] [CrossRef]
- Schulenberg, B.; Goodman, T.N.; Aggeler, R.; Capaldi, R.A.; Patton, W.F. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 2004, 25, 2526–2532. [Google Scholar] [CrossRef]
- Lenkowski, J.R.; Reed, J.M.; Deininger, L.; McLaughlin, K.A. Perturbation of organogenesis by the herbicide atrazine in the amphibian Xenopus laevis. Environ. Health Perspect. 2008, 116, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Fukamachi, K.; Han, B.S.; Kim, C.K.; Takasuka, N.; Matsuoka, Y.; Matsuda, E.; Yamasaki, T.; Tsuda, H. Possible enhancing effects of atrazine and nonylphenol on 7,12-dimethylbenz[a]anthraceneinduced mammary tumor development in human c-Ha-ras protooncogene transgenic rats. Cancer Sci. 2004, 95, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Dooley, G.P.; Reardon, K.F.; Prenni, J.E.; Tjalkens, R.B.; Legare, M.E.; Foradori, C.D.; Tessari, J.E.; Hanneman, W.H. Proteomic analysis of diaminochlorotriazine adducts in wistar rat pituitary glands and LbetaT2 rat pituitary cells. Chem. Res. Toxicol. 2008, 21, 844–851. [Google Scholar] [CrossRef]
- Pan, W.; da Graca, L.S.; Shao, Y.; Yin, Q.; Wu, H.; Jiang, X. PHAPI/pp32 suppresses tumorigenesis by stimulating apoptosis. J. Biol. Chem. 2009, 284, 6946–6954. [Google Scholar] [CrossRef] [PubMed]
- Walensky, L.D.; Coffey, D.; Chen, T.H.; Wu, T.C.; Pasternack, G.R. A novel Mr 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal1. Cancer Res. 1993, 53, 4720–4726. [Google Scholar] [PubMed]
- Li, Y.; Sun, Y.; Yang, J.; Wu, Y.; Yu, J.; Li, B. Age-dependent dopaminergic dysfunction following fetal exposure to atrazine in SD rats. Environ. Toxicol. Pharmacol. 2014, 37, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Kular, R.K.; Cvetanovic, M.; Siferd, S.; Kini, A.R.; Opal, P. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J. Biol. Chem. 2009, 284, 7783–7792. [Google Scholar] [CrossRef] [PubMed]
- Adegbola, O.; Pasternack, G.R. Phosphorylated retinoblastoma protein complexes with pp32 and inhibits pp32-mediated apoptosis. J. Biol. Chem. 2005, 280, 15497–15502. [Google Scholar] [CrossRef] [PubMed]
- Kadkol, S.S.; Brody, J.R.; Epstein, J.I.; Kuhajda, F.P.; Pasternack, G.R. Novel nuclear phosphoprotein pp32 is highly expressed in intermediate- and high-grade prostate cancer. Prostate 1998, 34, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Pechan, T.; Willett, K.L. Differential protein expression of peroxiredoxin I and II by benzo(a)pyrene and quercetin treatment in 22Rv1 and PrEC prostate cell lines. Toxicol. Appl. Pharmacol. 2007, 220, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xue, C.; Xue, D.; Zhao, J.; Gai, J.; Guo, N.; Xing, H. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS One 2013, 8, e69810. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Shigenaga, M.K.; Hagen, T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 1993, 90, 7915–7922. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, M.K.; Ames, B.N. Oxidants and mitogenesis as causes of mutation and cancer: The influence of diet. Basic Life Sci. 1993, 61, 419–436. [Google Scholar] [PubMed]
- Fatima, M.; Mandiki, S.N.; Douxfils, J.; Silvestre, F.; Coppe, P.; Kestemont, P. Combined effects of herbicides on biomarkers reflecting immune-endocrine interactions in goldfish. Immune and antioxidant effects. Aquat. Toxicol. 2007, 81, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Soo, E.T.; Yip, G.W.; Lwin, Z.M.; Kumar, S.D.; Bay, B.H. Heat shock proteins as novel therapeutic targets in cancer. In Vivo 2008, 22, 311–315. [Google Scholar]
- Edwards, D.P.; Adams, D.J.; McGuire, W.L. Estradiol stimulates synthesis of a major intracellular protein in a human breast cancer cell line (MCF-7). Breast Cancer Res. Treat. 1981, 1, 209–233. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, M.E.; Zhu, Y.; O’Neill, S. The 29-kDa proteins phosphorylated in thrombin-activated human platelets are forms of the estrogen receptor-related 27-kDa heat shock protein. Proc. Natl. Acad. Sci. USA 1991, 88, 11212–11216. [Google Scholar] [CrossRef] [PubMed]
- Dateki, M.; Kunitomo, M.; Yoshioka, K.; Yanai, K.; Nakasono, S.; Negishi, T. Adaptive gene regulation of pyruvate dehydrogenase kinase isoenzyme 4 in hepatotoxic chemical-induced liver injury and its stimulatory potential for DNA repair and cell proliferation. J. Recept. Signal Transduct. Res. 2011, 31, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Garaj-Vrhovac, V.; Zeljezic, D. Assessment of genome damage in a population of Croatian workers employed in pesticide production by chromosomal aberration analysis, micronucleus assay and Comet assay. J. Appl. Toxicol. 2002, 22, 249–255. [Google Scholar] [PubMed]
- Singh, M.; Kaur, P.; Sandhir, R.; Kiran, R. Protective effects of vitamin E against atrazine-induced genotoxicity in rats. Mutat. Res. 2008, 654, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Christl, T.J.; Pennington, P.; deLorenzo, M.; Karnaky, K.J., Jr.; Scott, G.I. Effect of multiple atrazine exposure profiles on hemocyte DNA integrity in the Eastern oyster (Crassostrea virginica). Bull. Environ. Contam. Toxicol. 2004, 73, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Shao, J.Z.; Xiang, L.X.; Chen, X.Y. Cytotoxic effects and apoptosis induction of atrazine in a grass carp (Ctenopharyngodon idellus) cell line. Environ. Toxicol. 2006, 21, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; An, S.; Henrich, V.C.; Sun, X.; Song, Q. Proteomic identification of PKC-mediated expression of 20E-induced protein in Drosophila melanogaster. J. Proteome Res. 2007, 6, 4478–4488. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.; Yang, J.; Song, Q. Atrazine Affects Phosphoprotein and Protein Expression in MCF-10A Human Breast Epithelial Cells. Int. J. Mol. Sci. 2014, 15, 17806-17826. https://doi.org/10.3390/ijms151017806
Huang P, Yang J, Song Q. Atrazine Affects Phosphoprotein and Protein Expression in MCF-10A Human Breast Epithelial Cells. International Journal of Molecular Sciences. 2014; 15(10):17806-17826. https://doi.org/10.3390/ijms151017806
Chicago/Turabian StyleHuang, Peixin, John Yang, and Qisheng Song. 2014. "Atrazine Affects Phosphoprotein and Protein Expression in MCF-10A Human Breast Epithelial Cells" International Journal of Molecular Sciences 15, no. 10: 17806-17826. https://doi.org/10.3390/ijms151017806




