A Functional Polymorphism in the 3'-UTR of PXR Interacts with Smoking to Increase Lung Cancer Risk in Southern and Eastern Chinese Smoker
Abstract
:1. Introduction
2. Results
2.1. PXR Genotypes and Lung Cancer Risk
| Genotypes | Case n (%) | Control a n (%) | p Value b | Crude OR (95% CI) | Adjusted OR c (95% CI) |
|---|---|---|---|---|---|
| Discovery set | |||||
| Total no. of subjects | 1056 | 1056 | |||
| rs3814055C>T | |||||
| CC | 693 (65.6) | 706 (66.9) | 0.836 | 1.00 | 1.00 |
| TC | 328 (31.1) | 316 (29.9) | 1.06 (0.88–1.27) | 1.06 (0.88–1.28) | |
| TT | 35 (3.3) | 34 (3.2) | 1.05 (0.65–1.70) | 1.05 (0.65–1.70) | |
| rs3732360C>T | |||||
| CC | 347 (32.9) | 346 (32.8) | 0.758 | 1.00 | 1.00 |
| TC | 520 (49.2) | 533 (50.5) | 0.97 (0.80–1.18) | 0.97 (0.80–1.18) | |
| TT | 189 (17.9) | 177 (16.7) | 1.07 (0.83–1.37) | 1.07 (0.83–1.37) | |
| rs3814058C>T | |||||
| CC | 315 (29.8) | 365 (34.6) | 0.033 | 1.00 | 1.00 |
| TC | 505 (47.8) | 491 (46.5) | 1.19 (0.98–1.45) | 1.19 (0.98–1.45) | |
| TT | 236 (22.4) | 200 (18.9) | 1.37 (1.07–1.74) | 1.36 (1.07–1.73) | |
| TC + TT | 741 (70.2) | 691 (65.4) | 1.24 (1.04–1.49) | 1.24 (1.03–1.49) | |
| Validation set | |||||
| Total no. of subjects | 503 | 623 | |||
| rs3814058C>T | |||||
| CC | 122 (24.2) | 185 (29.7) | 0.093 | 1.00 | 1.00 |
| TC | 254 (50.5) | 303 (48.6) | 1.27 (0.96–1.68) | 1.28 (0.96–1.70) | |
| TT | 127 (25.3) | 135 (21.7) | 1.43 (1.02–1.99) | 1.47 (1.05–2.05) | |
| TC + TT | 381 (75.8) | 438 (70.3) | 1.32 (1.01–1.72) | 1.33 (1.02–1.75) | |
| Merged set | |||||
| Total no. of subjects | 1559 | 1679 | |||
| rs3814058C>T | |||||
| CC | 437 (28.0) | 550 (32.8) | 0.006 | 1.00 | 1.00 |
| TC | 759 (48.7) | 794 (47.2) | 1.20 (1.03–1.41) | 1.20 (1.02–1.41) | |
| TT | 363 (23.3) | 335 (20.0) | 1.36 (1.12–1.66) | 1.38 (1.13–1.67) | |
| Dominant model | |||||
| CC | 437 (28.0) | 550 (32.8) | 1.00 | 1.00 | |
| TC + TT | 1,122 (72.0) | 1,129 (67.2) | 1.25 (1.08–1.45) | 1.25 (1.08–1.45) | |
2.2. Stratification Analysis
| Variables | Cases (n = 1559) | Controls (n = 1679) | Adjusted OR a (95% CI) | p b | ||
|---|---|---|---|---|---|---|
| TG + TT n (%) | CC n (%) | TG + TT n (%) | CC n (%) | TC + TT vs. CC | ||
| Age (years) | ||||||
| ≤60 | 586 (72.4) | 223 (27.6) | 595 (67.8) | 282 (32.2) | 1.24 (1.01–1.54) | 0.884 |
| >60 | 536 (71.5) | 214 (28.5) | 534 (66.6) | 268 (33.4) | 1.24 (1.00–1.54) | |
| Sex | ||||||
| Male | 793 (72.7) | 298 (27.3) | 787(66.4) | 398 (33.6) | 1.35 (1.13–1.62) | 0.107 |
| Female | 329 (70.3) | 139 (29.7) | 342(69.2) | 152 (30.8) | 1.03 (0.78–1.35) | |
| Smoking status | ||||||
| Current | 371 (72.4) | 141 (27.6) | 354 (66.5) | 178 (33.5) | 1.33 (1.02–1.73) | 0.023 |
| Former | 241 (77.2) | 71 (22.8) | 150 (64.4) | 83 (35.6) | 1.92 (1.31–2.81) | |
| Never | 510 (69.4) | 225 (30.6) | 625 (68.4) | 289 (31.6) | 1.06 (0.85–1.31) | |
| Pack-years smoked | ||||||
| ≥20 | 457 (73.2) | 167 (26.8) | 159 (33.2) | 320 (66.8) | 1.34 (1.04–1.75) | 0.019 |
| <20 | 155 (77.5) | 45 (22.5) | 119 (32.5) | 247 (67.5) | 1.93 (1.28–2.93) | |
| 0 | 510 (69.4) | 225 (30.6) | 272 (32.6) | 562 (67.4) | 1.10 (0.89–1.36) | |
| Drinking status | ||||||
| Ever | 218 (74.4) | 75 (25.6) | 231 (67.5) | 111 (32.5) | 1.48 (1.03–2.11) | 0.427 |
| Never | 904 (71.4) | 362 (28.6) | 898 (67.2) | 439 (32.8) | 1.21 (1.03–1.43) | |
| Family history of cancer | ||||||
| Yes | 91 (70.5) | 38 (29.5) | 98 (66.7) | 49 (33.3) | 1.15 (0.68–1.94) | 0.821 |
| No | 1031 (72.1) | 399 (27.9) | 1031 (67.3) | 501 (32.7) | 1.25 (1.07–1.47) | |
| Family history of lung cancer | ||||||
| Yes | 40 (76.9) | 12 (23.1) | 28 (65.1) | 15 (34.9) | 1.82 (0.67–4.91) | 0.082 |
| No | 1082 (71.8) | 425 (28.2) | 1101 (67.3) | 535 (32.7) | 1.23 (1.06–1.43) | |
| Histological types | ||||||
| Adenocarcinoma | 431 (70.1) | 184 (29.9) | 1129 (67.2) | 550 (32.8) | 1.14 (0.93–1.40) | |
| Squamous cell carcinoma | 385 (73.1) | 142 (26.9) | 1.31 (1.05–1.63) | |||
| Large cell carcinoma | 45 (68.2) | 21 (31.8) | 1.07 (0.63–1.81) | |||
| Small cell lung cancer | 152 (78.8) | 41 (21.2) | 1.83 (1.28–2.63) | |||
| Other carcinomas c | 109 (69.0) | 49 (31.0) | 1.08 (0.76–1.54) | |||
| Stages | ||||||
| I | 136 (68.0) | 64 (32.0) | 1129 (67.2) | 550 (32.8) | 1.04 (0.76–1.42) | |
| II | 105 (71.4) | 42 (28.6) | 1.22 (0.84–1.77) | |||
| III | 357 (72.9) | 133 (27.1) | 1.31 (1.05–1.64) | |||
| IV | 524 (72.6) | 198 (27.4) | 1.30 (1.07–1.57) | |||
2.3. Association between the rs3814058C>T Genotypes and mRNA Levels of PXR Gene
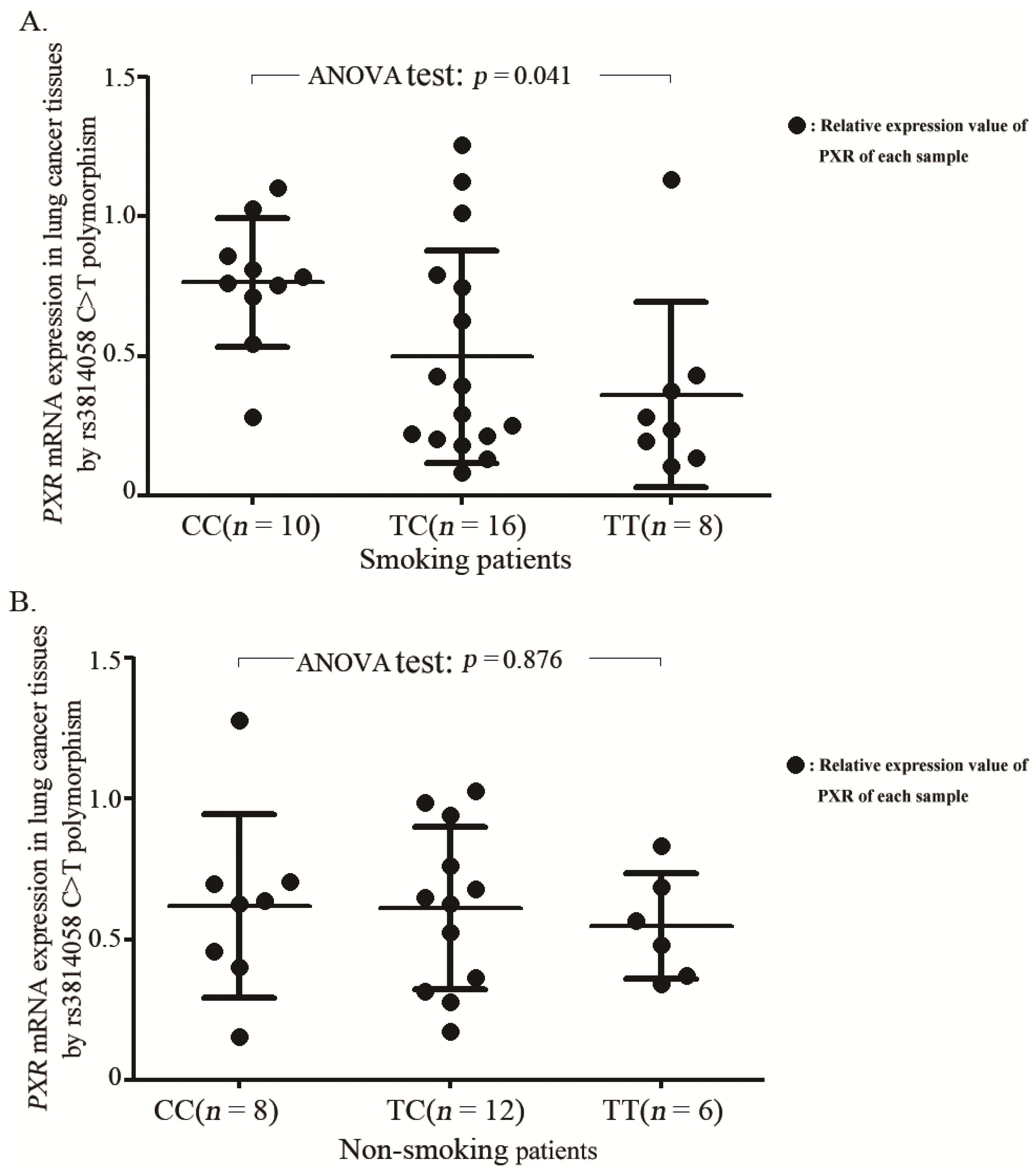
3. Discussion
4. Experimental Section
4.1. Study Subjects
4.2. Single Nucleotide Polymorphism (SNP) Selection and Genotyping
4.3. PXR mRNA Expression Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, W.; Zheng, R.; Zhang, S.; Zhao, P.; Zeng, H.; Zou, X.; He, J. Annual report on status of cancer in China, 2010. Chin. J. Cancer Res. 2014, 26, 48–58. [Google Scholar] [PubMed]
- Gupta, A.; Srivastava, S.; Prasad, R.; Natu, S.M.; Mittal, B.; Negi, M.P.; Srivastava, A.N. Smoking intensity, oxidative stress and chemotherapy in nonsmall cell lung cancer: A correlated prognostic study. Biosci. Trends 2009, 3, 191–199. [Google Scholar] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoke and involuntary smoking. IARC Monogr. Eval. Carcinog. Risks Hum. 2004, 83, 1–1438. [Google Scholar]
- Czerwinski, M.; McLemore, T.L.; Gelboin, H.V.; Gonzalez, F.J. Quantification of CYP2b7, CYP4b1, and CYPOR messenger RNAs in normal human lung and lung tumors. Cancer Res. 1994, 54, 1085–1091. [Google Scholar] [PubMed]
- Denissenko, M.F.; Pao, A.; Tang, M.; Pfeifer, G.P. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in p53. Science 1996, 274, 430–432. [Google Scholar] [PubMed]
- Van de Winkel, A.; Menke, V.; Capello, A.; Moons, L.M.; Pot, R.G.; van Dekken, H.; Siersema, P.D.; Kusters, J.G.; van der Laan, L.J.; Kuipers, E.J. Expression, localization and polymorphisms of the nuclear receptor PXR in barrett’s esophagus and esophageal adenocarcinoma. BMC Gastroenterol. 2011, 11, 108. [Google Scholar] [PubMed]
- Kotta-Loizou, I.; Patsouris, E.; Theocharis, S. Pregnane X receptor polymorphisms associated with human diseases. Expert Opin. Ther. Targets 2013, 17, 1167–1177. [Google Scholar] [PubMed]
- Sookoian, S.; Castano, G.O.; Burgueno, A.L.; Gianotti, T.F.; Rosselli, M.S.; Pirola, C.J. The nuclear receptor PXR gene variants are associated with liver injury in nonalcoholic fatty liver disease. Pharmacogenet. Genomics 2010, 20, 1–8. [Google Scholar] [PubMed]
- Dring, M.M.; Goulding, C.A.; Trimble, V.I.; Keegan, D.; Ryan, A.W.; Brophy, K.M.; Smyth, C.M.; Keeling, P.W.; O’Donoghue, D.; O’Sullivan, M.; et al. The pregnane X receptor locus is associated with susceptibility to inflammatory bowel disease. Gastroenterology 2006, 130, 341–348. [Google Scholar]
- Kliewer, S.A.; Moore, J.T.; Wade, L.; Staudinger, J.L.; Watson, M.A.; Jones, S.A.; McKee, D.D.; Oliver, B.B.; Willson, T.M.; Zetterstrom, R.H.; et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998, 92, 73–82. [Google Scholar]
- Giguere, V. Orphan nuclear receptors: From gene to function. Endocr. Rev. 1999, 20, 689–725. [Google Scholar] [PubMed]
- Gonzalez, F.J. Human cytochromes p450: Problems and prospects. Trends Pharmacol. Sci. 1992, 13, 346–352. [Google Scholar] [PubMed]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes p450. Lancet 2002, 360, 1155–1162. [Google Scholar] [PubMed]
- Goodwin, B.; Moore, L.B.; Stoltz, C.M.; McKee, D.D.; Kliewer, S.A. Regulation of the human CYP2b6 gene by the nuclear pregnane X receptor. Mol. Pharmacol. 2001, 60, 427–431. [Google Scholar] [PubMed]
- Hodgson, E.; Rose, R.L. The importance of cytochrome p450 2b6 in the human metabolism of environmental chemicals. Pharmacol. Ther. 2007, 113, 420–428. [Google Scholar] [PubMed]
- Kumagai, T.; Suzuki, H.; Sasaki, T.; Sakaguchi, S.; Miyairi, S.; Yamazoe, Y.; Nagata, K. Polycyclic aromatic hydrocarbons activate CYP3a4 gene transcription through human pregnane X receptor. Drug Metab. Pharmacokinet. 2012, 27, 200–206. [Google Scholar] [PubMed]
- Oyama, T.; Uramoto, H.; Kagawa, N.; Yoshimatsu, T.; Osaki, T.; Nakanishi, R.; Nagaya, H.; Kaneko, K.; Muto, M.; Kawamoto, T.; et al. Cytochrome p450 in non-small cell lung cancer related to exogenous chemical metabolism. Front. Biosci. 2012, 4, 1539–1546. [Google Scholar]
- Gonzalez, F.J.; Gelboin, H.V. Role of human cytochromes p450 in the metabolic activation of chemical carcinogens and toxins. Drug Metab. Rev. 1994, 26, 165–183. [Google Scholar] [PubMed]
- Oyama, T.; Sugio, K.; Isse, T.; Matsumoto, A.; Nose, N.; Uramoto, H.; Nozoe, T.; Morita, M.; Kagawa, N.; Osaki, T.; et al. Expression of cytochrome p450 in non-small cell lung cancer. Front. Biosci. 2008, 13, 5787–5793. [Google Scholar] [PubMed]
- Chirulli, V.; Longo, V.; Marini, S.; Mazzaccaro, A.; Fiorio, R.; Gervasi, P.G. Car and PXR expression and inducibility of CYP2b and CYP3a activities in rat and rabbit lungs. Life Sci. 2005, 76, 2535–2546. [Google Scholar] [PubMed]
- Kliewer, S.A. The nuclear pregnane X receptor regulates xenobiotic detoxification. J. Nutr. 2003, 133, 2444S–2447S. [Google Scholar] [PubMed]
- Dai, G.; He, L.; Bu, P.; Wan, Y.J. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology 2008, 47, 1277–1287. [Google Scholar] [PubMed]
- Gupta, D.; Venkatesh, M.; Wang, H.; Kim, S.; Sinz, M.; Goldberg, G.L.; Whitney, K.; Longley, C.; Mani, S. Expanding the roles for pregnane X receptor in cancer: Proliferation and drug resistance in ovarian cancer. Clin. Cancer Res. 2008, 14, 5332–5340. [Google Scholar] [PubMed]
- Zucchini, N.; de Sousa, G.; Bailly-Maitre, B.; Gugenheim, J.; Bars, R.; Lemaire, G.; Rahmani, R. Regulation of Bcl-2 and Bcl-xl anti-apoptotic protein expression by nuclear receptor PXR in primary cultures of human and rat hepatocytes. Biochim. Biophys. Acta 2005, 1745, 48–58. [Google Scholar] [PubMed]
- Huang, R.; Murry, D.J.; Kolwankar, D.; Hall, S.D.; Foster, D.R. Vincristine transcriptional regulation of efflux drug transporters in carcinoma cell lines. Biochem. Pharmacol. 2006, 71, 1695–1704. [Google Scholar] [PubMed]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar]
- Robbins, D.; Chen, T. Tissue-specific regulation of pregnane X receptor in cancer development and therapy. Cell Biosci. 2014, 4, 17. [Google Scholar] [PubMed]
- Miki, Y.; Suzuki, T.; Kitada, K.; Yabuki, N.; Shibuya, R.; Moriya, T.; Ishida, T.; Ohuchi, N.; Blumberg, B.; Sasano, H. Expression of the steroid and xenobiotic receptor and its possible target gene, organic anion transporting polypeptide-A, in human breast carcinoma. Cancer Res. 2006, 66, 535–542. [Google Scholar] [PubMed]
- Ouyang, N.; Ke, S.; Eagleton, N.; Xie, Y.; Chen, G.; Laffins, B.; Yao, H.; Zhou, B.; Tian, Y. Pregnane X receptor suppresses proliferation and tumourigenicity of colon cancer cells. Br. J. Cancer 2010, 102, 1753–1761. [Google Scholar] [PubMed]
- Snpinfo Web Server. Available online: http://snpinfo.niehs.nih.gov/ (accessed on 18 June 2014).
- Dong, J.; Hu, Z.; Wu, C.; Guo, H.; Zhou, B.; Lv, J.; Lu, D.; Chen, K.; Shi, Y.; Chu, M.; et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the chinese population. Nat. Genet. 2012, 44, 895–899. [Google Scholar]
- Liu, B.; Chen, D.; Yang, L.; Li, Y.; Ling, X.; Liu, L.; Ji, W.; Wei, Y.; Wang, J.; Wei, Q.; et al. A functional variant (–1304t>g) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis 2010, 31, 1405–1411. [Google Scholar]
- Yang, L.; Li, Y.; Cheng, M.; Huang, D.; Zheng, J.; Liu, B.; Ling, X.; Li, Q.; Zhang, X.; Ji, W.; et al. A functional polymorphism at microRNA-629-binding site in the 3'-untranslated region of NBS1 gene confers an increased risk of lung cancer in southern and eastern Chinese population. Carcinogenesis 2012, 33, 338–347. [Google Scholar]
- HapMap Database. Available online: http://hapmap.ncbi.nlm.nih.gov/index.html.en (accessed on 20 October 2013).
- Lu, J.; Yang, L.; Zhao, H.; Liu, B.; Li, Y.; Wu, H.; Li, Q.; Zeng, B.; Wang, Y.; Ji, W.; et al. The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum. Mutat. 2011, 32, 1299–1308. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative Ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]
- Dupont, W.D.; Plummer, W.D., Jr. Power and sample size calculations for studies involving linear regression. Control Clin. Trials 1998, 19, 589–601. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Qiu, F.; Lu, X.; Li, Y.; Fang, W.; Zhang, L.; Zhou, Y.; Yang, L.; Lu, J. A Functional Polymorphism in the 3'-UTR of PXR Interacts with Smoking to Increase Lung Cancer Risk in Southern and Eastern Chinese Smoker. Int. J. Mol. Sci. 2014, 15, 17457-17468. https://doi.org/10.3390/ijms151017457
Zhang L, Qiu F, Lu X, Li Y, Fang W, Zhang L, Zhou Y, Yang L, Lu J. A Functional Polymorphism in the 3'-UTR of PXR Interacts with Smoking to Increase Lung Cancer Risk in Southern and Eastern Chinese Smoker. International Journal of Molecular Sciences. 2014; 15(10):17457-17468. https://doi.org/10.3390/ijms151017457
Chicago/Turabian StyleZhang, Lisha, Fuman Qiu, Xiaoxiao Lu, Yinyan Li, Wenxiang Fang, Lan Zhang, Yifeng Zhou, Lei Yang, and Jiachun Lu. 2014. "A Functional Polymorphism in the 3'-UTR of PXR Interacts with Smoking to Increase Lung Cancer Risk in Southern and Eastern Chinese Smoker" International Journal of Molecular Sciences 15, no. 10: 17457-17468. https://doi.org/10.3390/ijms151017457




