Characterization of an Invertebrate-Type Dopamine Receptor of the American Cockroach, Periplaneta americana
Abstract
:1. Introduction
2. Results
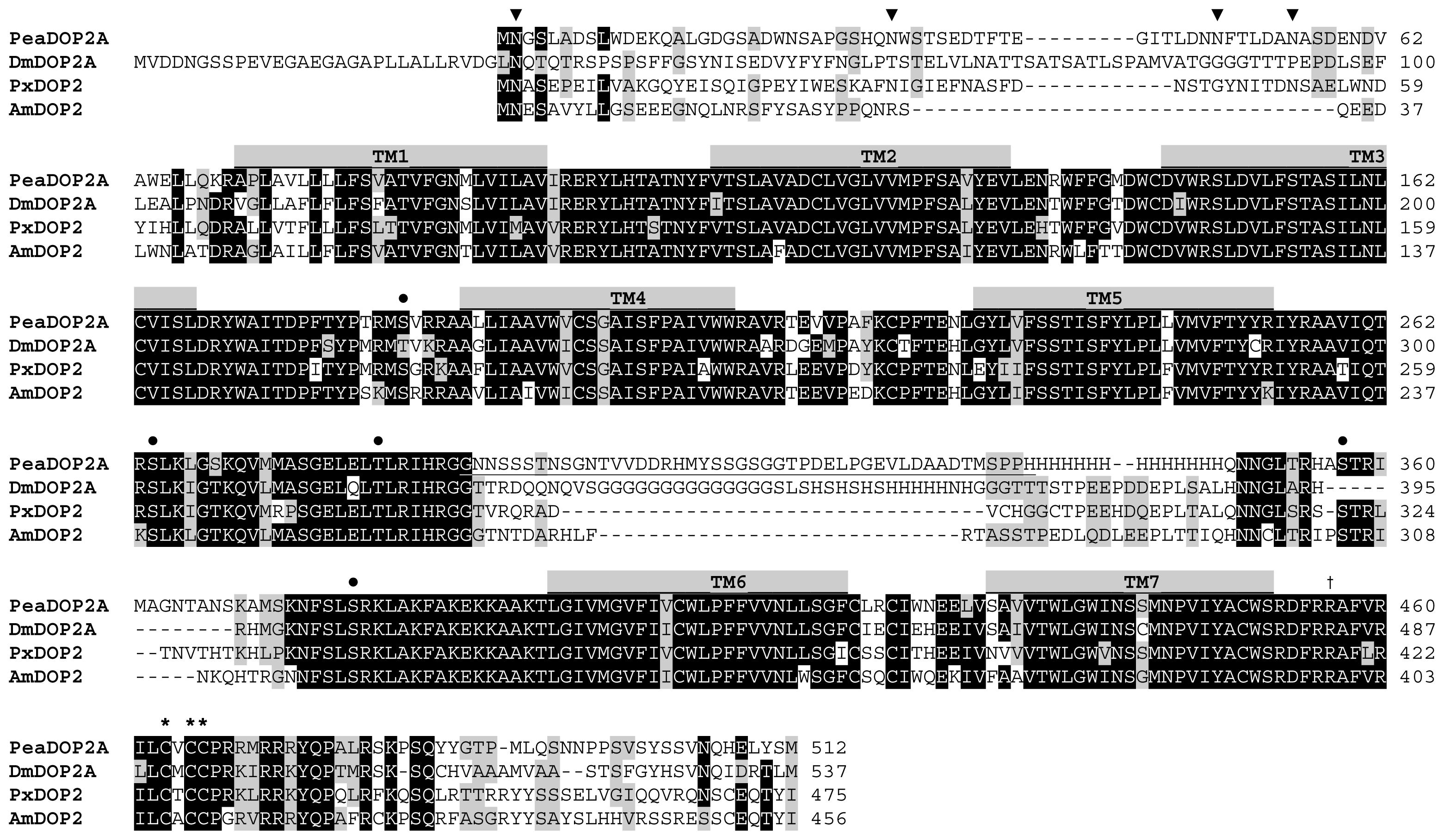
2.1. Molecular Cloning and Sequence Analysis of a Dopamine Receptor from P. americana
2.2. Tissue Distribution of Peadop2A and Peadop2B mRNA
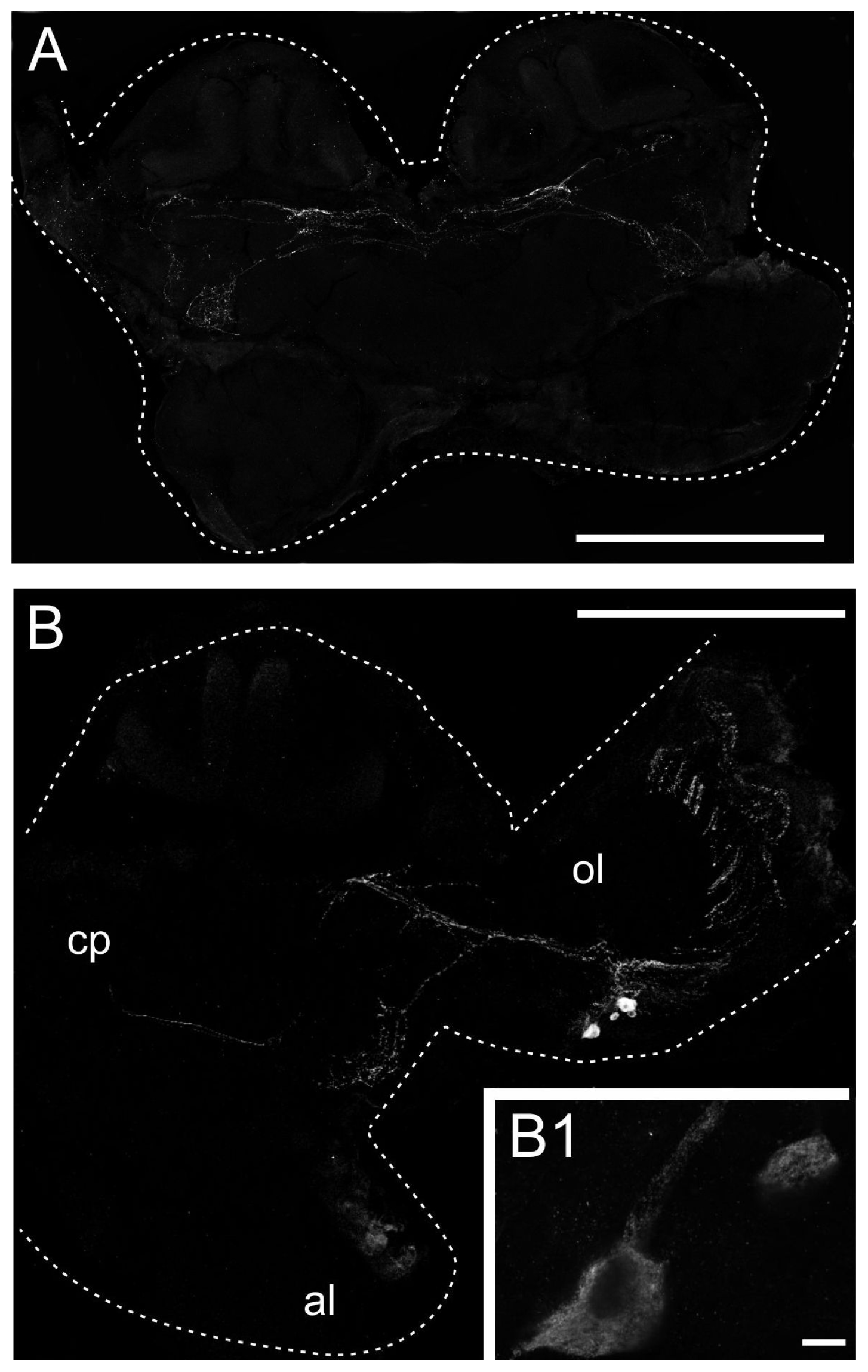
2.3. Generation of an Anti-PeaDOP2 Antibody and Immunohistochemical Localization of PeaDOP2A/B Receptors
2.4. Functional Analyses of PeaDOP2A/B Receptors in HEK 293 Cells
3. Discussion
3.1. Receptor Variants Occur by Alternative Splicing of the Peadop2 Gene
3.2. Structural Characteristics of the PeaDOP2 Receptor
3.3. PeaDOP2A but Not PeaDOP2B Is a Functional Dopamine Receptor
3.4. Pharmacological Properties of the PeaDOP2A Receptor
3.5. Distribution of the PeaDOP2 Receptor in the Nervous System
3.6. Possible Function of the PeaDOP2A Receptor in Saliva Generation and Secretion
4. Experimental Section
4.1. Receptor Ligands
4.2. Cloning of Peadop2 cDNA
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. RT-PCR Amplification of Peadop2A and Peadop2B Fragments
4.5. Antibody Production and Purification
4.6. Western Blot Analysis
4.7. Immunofluorescence Staining of Brain Sections
4.8. Construction of pcPeadop2A/B-HA Expression Vector
4.9. Functional Expression of the PeaDOP2A/B-HA Receptor
4.10. Functional Characterization of PeaDOP2A and PeaDOP2B Receptors
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Huber, I.; Masler, E.P.; Rao, B.R. Cockroaches as Models for Neurobiology: Applications in Biomedical Research, 1st ed; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Page, T.L. Serotonin phase-shifts the circadian rhythm of locomotor activity in the cockroach. J. Biol. Rhythms 1987, 2, 23–34. [Google Scholar]
- Matsui, T.; Matsumoto, T.; Ichihara, N.; Sakai, T.; Satake, H.; Watari, Y.; Takeda, M. The pars intercerebralis as a modulator of locomotor rhythms and feeding in the American cockroach Periplaneta americana. Physiol. Behav 2009, 96, 548–556. [Google Scholar]
- Helfrich-Förster, C. The circadian clock in the brain: A structural and functional comparison between mammals and insects. J. Comp. Physiol. A 2004, 190, 601–613. [Google Scholar]
- Camhi, J.M. Escape behavior in the cockroach: Distributed neural processing. Experientia 1988, 44, 401–408. [Google Scholar]
- Predel, R.; Eckert, M. Neurosecretion: Peptidergic systems in insects. Naturwissenschaften 2000, 87, 343–350. [Google Scholar]
- Just, F.; Walz, B. The effects of serotonin and dopamine on salivary secretion by isolated cockroach salivary glands. J. Exp. Biol 1996, 199, 407–413. [Google Scholar]
- Marg, S.; Walz, B.; Blenau, W. The effects of dopamine receptor agonists and antagonists on the secretory rate of cockroach (Periplaneta americana) salivary glands. J. Insect Physiol 2004, 50, 821–830. [Google Scholar]
- Rietdorf, K.; Lang, I.; Walz, B. Saliva secretion and ionic composition of saliva in the cockroach Periplaneta americana after serotonin and dopamine stimulation, and effects of ouabain and bumetamide. J. Insect Physiol 2003, 49, 205–215. [Google Scholar]
- Troppmann, B.; Walz, B.; Blenau, W. Pharmacology of serotonin-induced salivary secretion in Periplaneta americana. J. Insect Physiol 2007, 53, 774–781. [Google Scholar]
- House, C.R.; Ginsborg, B.L. Salivary Gland. In Comprehensive Insect Physiology, Biochemistry and Pharmacology, 1st ed; Kerkut, G.A., Gilbert, L.I., Eds.; Pergamon Press: Oxford, UK, 1985; Volume 11, pp. 195–224. [Google Scholar]
- Walz, B.; Baumann, O.; Krach, C.; Baumann, A.; Blenau, W. The aminergic control of cockroach salivary glands. Arch. Insect Biochem. Physiol 2006, 62, 141–152. [Google Scholar]
- Watanabe, H.; Kobayashi, Y.; Sakura, M.; Matsumoto, Y.; Mizunami, M. Classical olfactory conditioning in the cockroach Periplaneta americana. Zool. Sci 2003, 20, 1447–1454. [Google Scholar]
- Kwon, H.W.; Lent, D.D.; Strausfeld, N.J. Spatial learning in the restrained American cockroach Periplaneta americana. J. Exp. Biol 2004, 207, 377–383. [Google Scholar]
- Lent, D.D.; Kwon, H.W. Antennal movements reveal associative learning in the American cockroach Periplaneta americana. J. Exp. Biol 2004, 207, 369–375. [Google Scholar]
- Watanabe, H.; Mizunami, M. Pavlov’s cockroach: Classical conditioning of salivation in an insect. PLoS One 2007, 2, e529. [Google Scholar]
- Matsumoto, C.S.; Matsumoto, Y.; Watanabe, H.; Nishino, H.; Mizunami, M. Context-dependent olfactory learning monitored by activities of salivary neurons in cockroaches. Neurobiol. Learn. Mem 2012, 97, 30–36. [Google Scholar]
- Blenau, W.; Baumann, A. Molecular and pharmacological properties of insect biogenic amine receptors: Lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Physiol 2001, 48, 13–38. [Google Scholar]
- Mustard, J.A.; Beggs, K.T.; Mercer, A.R. Molecular biology of the invertebrate dopamine receptors. Arch. Insect Biochem. Physiol 2005, 59, 103–117. [Google Scholar]
- Evans, P.D. Biogenic amines in the insect nervous system. Adv. Insect Physiol 1980, 15, 317–473. [Google Scholar]
- Downer, R.G.H. Octopamine, Dopamine, and 5-Hydroxytryptamine in the Cockroach Nervous System. In Cockroaches as Models for Neurobiology: Applications in Biomedical Research, 1st ed; Huber, I., Masler, E.P., Rao, B.R., Eds.; CRC Press: Boca Raton, FL, USA, 1990; Volume 2, pp. 103–124. [Google Scholar]
- Distler, P. Synaptic connections of dopamine-immunoreactive neurons in the antennal lobes of Periplaneta americana. Colocalization with GABA-like immunoreactivity. Histochemistry 1990, 93, 401–408. [Google Scholar]
- Milton, G.W.; Verhaert, P.D.; Downer, R.G. Immunofluorescent localization of dopamine-like and leucine-enkephalin-like neurons in the supraoesophageal ganglia of the American cockroach Periplaneta americana. Tissue Cell 1991, 23, 331–340. [Google Scholar]
- Granholm, A.C.; Price, M.L.; Owen, M.D. Tyrosine hydroxylase in the cerebral ganglia of the American cockroach (Periplaneta americana L.): An immunohistochemical study. Cell Tissue Res 1995, 282, 49–57. [Google Scholar]
- Budnik, V.; White, K. Catecholamine-containing neurons in Drosophila melanogaster: Distribution and development. J. Comp. Neurol 1988, 268, 400–413. [Google Scholar]
- Schäfer, S.; Rehder, V. Dopamine-like immunoreactivity in the brain and suboesophageal ganglion of the honeybee. J. Comp. Neurol 1989, 280, 43–58. [Google Scholar]
- Blenau, W.; Schmidt, M.; Faensen, D.; Schürmann, F.W. Neurons with dopamine-like immunoreactivity target the mushroom body Kenyon cell somata in the brain of some hymenopteran insects. Int. J. Insect Morphol. Embryol 1999, 28, 203–210. [Google Scholar]
- Wendt, B.; Homberg, U. Immunocytochemistry of dopamine in the brain of the locust Schistocerca gregaria. J. Comp. Neurol 1992, 321, 387–403. [Google Scholar]
- Gifford, A.N.; Nicholson, R.A.; Pitman, R.M. The dopamine and 5-hydroxytryptamine content of locust and cockroach salivary neurones. J. Exp. Biol 1991, 161, 405–414. [Google Scholar]
- Baumann, O.; Dames, P.; Kühnel, D.; Walz, B. Distribution of serotonergic and dopaminergic nerve fibers in the salivary gland complex of the cockroach Periplaneta americana. BMC Physiol 2002, 2, 9. [Google Scholar]
- Ali, D. The aminergic and peptidergic innervation of insect salivary glands. J. Exp. Biol 1997, 200, 1941–1949. [Google Scholar]
- Schwaerzel, M.; Monastirioti, M.; Scholz, H.; Friggi-Grelin, F.; Birman, S.; Heisenberg, M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci 2003, 23, 10495–10502. [Google Scholar]
- Andretic, R.; van Swinderen, B.; Greenspan, R.J. Dopaminergic modulation of arousal in Drosophila. Curr. Biol 2005, 15, 1165–1175. [Google Scholar]
- Kume, K.; Kume, S.; Park, S.K.; Hirsh, J.; Jackson, F.R. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci 2005, 25, 7377–7384. [Google Scholar]
- Neckameyer, W.S.; Weinstein, J.S. Stress affects dopaminergic signaling pathways in Drosophila melanogaster. Stress 2005, 8, 117–131. [Google Scholar]
- Chang, H.Y.; Grygoruk, A.; Brooks, E.S.; Ackerson, L.C.; Maidment, N.T.; Bainton, R.J.; Krantz, D.E. Overexpression of the Drosophila vesicular monoamine transporter increases motor activity and courtship but decreases the behavioral response to cocaine. Mol. Psychiatry 2006, 11, 99–113. [Google Scholar]
- Draper, I.; Kurshan, P.T.; McBride, E.; Jackson, F.R.; Kopin, A.S. Locomotor activity is regulated by D2-like receptors in Drosophila: An anatomic and functional analysis. Dev. Neurobiol 2007, 67, 378–393. [Google Scholar]
- Bayersdorfer, F.; Voigt, A.; Schneuwly, S.; Botella, J.A. Dopamine-dependent neurodegeneration in Drosophila models of familial and sporadic Parkinson’s disease. Neurobiol. Dis 2010, 40, 113–119. [Google Scholar]
- Hirsh, J.; Riemensperger, T.; Coulom, H.; Iché, M.; Coupar, J.; Birman, S. Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr. Biol 2010, 20, 209–214. [Google Scholar]
- Riemensperger, T.; Isabel, G.; Coulom, H.; Neuser, K.; Seugnet, L.; Kume, K.; Iché-Torres, M.; Cassar, M.; Strauss, R.; Preat, T.; et al. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc. Natl. Acad. Sci. USA 2011, 108, 834–839. [Google Scholar]
- Neckameyer, W.S. Multiple roles for dopamine in Drosophila development. Dev. Biol 1996, 176, 209–219. [Google Scholar]
- Cooper, R.L.; Neckameyer, W.S. Dopaminergic modulation of motor neuron activity and neuromuscular function in Drosophila melanogaster. Comp. Biochem. Physiol. B 1999, 122, 199–210. [Google Scholar]
- Keating, C.; Orchard, I. Dopamine induces hyperpolarization of locust salivary gland acinar cells via D1-like receptors. J. Insect Physiol 2001, 47, 667–673. [Google Scholar]
- Keating, C.; Orchard, I. The effects of dopamine agonists and antagonists on the secretory responses in the salivary glands of the locust (Locusta migratoria). J. Insect Physiol 2004, 50, 17–23. [Google Scholar]
- Granger, N.A.; Sturgis, S.L.; Ebersohl, R.; Geng, C.; Sparks, T.C. Dopaminergic control of corpora allata activity in the larval tobacco hornworm Manduca sexta. Arch. Insect Biochem. Physiol 1996, 32, 449–466. [Google Scholar]
- Gruntenko, N.E.; Karpova, E.K.; Alekseev, A.A.; Chentsova, N.A.; Saprykina, Z.V.; Bownes, M.; Rauschenbach, I.Y. Effects of dopamine on juvenile hormone metabolism and fitness in Drosophila virilis. J. Insect Physiol 2005, 51, 959–968. [Google Scholar]
- Missale, C.; Nash, S.R.; Robinson, S.W.; Jaber, M.; Caron, M.G. Dopamine receptors: From structure to function. Physiol. Rev 1998, 78, 189–225. [Google Scholar]
- Neve, K.A.; Seamans, J.K.; Trantham-Davidson, H. Dopamine receptor signaling. J. Recept. Signal Transduct. Res 2004, 24, 165–205. [Google Scholar]
- Gotzes, F.; Balfanz, S.; Baumann, A. Primary structure and functional characterization of a Drosophila dopamine receptor with high homology to human D1/5 receptors. Recept. Channels 1994, 2, 131–141. [Google Scholar]
- Hearn, M.G.; Ren, Y.; McBride, E.W.; Reveillaud, I.; Beinborn, M.; Kopin, A.S. A Drosophila dopamine 2-like receptor: Molecular characterization and identification of multiple alternatively spliced variants. Proc. Natl. Acad. Sci. USA 2002, 99, 14554–14559. [Google Scholar]
- Blenau, W.; Erber, J.; Baumann, A. Characterization of a dopamine D1 receptor from Apis mellifera: Cloning, functional expression, pharmacology, and mRNA localization in the brain. J. Neurochem 1998, 70, 15–23. [Google Scholar]
- Beggs, K.T.; Hamilton, I.S.; Kurshan, P.T.; Mustard, J.A.; Mercer, A.R. Characterization of a D2-like dopamine receptor (AmDOP3) in honey bee Apis mellifera. Insect Biochem. Mol. Biol 2005, 35, 873–882. [Google Scholar]
- Feng, G.; Hannan, F.; Reale, V.; Hon, Y.Y.; Kousky, C.T.; Evans, P.D.; Hall, L.M. Cloning and functional characterization of a novel dopamine receptor from Drosophila melanogaster. J. Neurosci 1996, 16, 3925–3933. [Google Scholar]
- Han, K.A.; Millar, N.S.; Grotewiel, M.S.; Davis, R.L. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron 1996, 16, 1127–1135. [Google Scholar]
- Humphries, M.A.; Mustard, J.A.; Hunter, S.J.; Mercer, A.; Ward, V.; Ebert, P.R. Invertebrate D2 type dopamine receptor exhibits age-based plasticity of expression in the mushroom bodies of the honeybee brain. J. Neurobiol 2003, 55, 315–330. [Google Scholar]
- Mustard, J.A.; Blenau, W.; Hamilton, I.S.; Ward, V.K.; Ebert, P.R.; Mercer, A.R. Analysis of two D1-like dopamine receptors from the honey bee Apis mellifera reveals agonist-independent activity. Brain Res. Mol. Brain Res 2003, 113, 67–77. [Google Scholar]
- Evans, P.D.; Maqueira, B. Insect octopamine receptors: A new classification scheme based on studies of cloned Drosophila G-protein coupled receptors. Invert. Neurosci 2005, 5, 111–118. [Google Scholar]
- Beggs, K.T.; Tyndall, J.D.; Mercer, A.R. Honey bee dopamine and octopamine receptors linked to intracellular calcium signaling have a close phylogenetic and pharmacological relationship. PLoS One 2011, 6, e26809. [Google Scholar]
- Bischof, L.J.; Enan, E.E. Cloning, expression and functional analysis of an octopamine receptor from Periplaneta americana. Insect Biochem. Mol. Biol 2004, 34, 511–521. [Google Scholar]
- Rotte, C.; Krach, C.; Balfanz, S.; Baumann, A.; Walz, B.; Blenau, W. Molecular characterization and localization of the first tyramine receptor of the American cockroach (Periplaneta americana). Neuroscience 2009, 162, 1120–1133. [Google Scholar]
- Troppmann, B.; Balfanz, S.; Baumann, A.; Blenau, W. Inverse agonist and neutral antagonist actions of synthetic compounds at an insect 5-HT1 receptor. Br. J. Pharmacol 2010, 159, 1450–1462. [Google Scholar]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol 2004, 338, 1027–1036. [Google Scholar]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci 1995, 25, 366–428. [Google Scholar]
- Strader, C.; Fong, T.; Graziano, M.; Tota, M. The family of G-protein-coupled receptors. FASEB J 1995, 9, 745–754. [Google Scholar]
- Kroeze, W.K.; Kristiansen, K.; Roth, B.L. Molecular biology of 5-HT receptors-structure and function at the molecular level. Curr. Top. Med. Chem 2002, 2, 507–528. [Google Scholar]
- Ono, H.; Yoshikawa, H. Identification of amine receptors from a swallowtail butterfly, Papilio xuthus L.: Cloning and mRNA localization in foreleg chemosensory organ for recognition of host plants. Insect Biochem. Mol. Biol 2004, 34, 1247–1256. [Google Scholar]
- Gerber, S.; Krasky, A.; Rohwer, A.; Lindauer, S.; Closs, E.; Rognan, D.; Gunkel, N.; Selzer, P.M.; Wolf, C. Identification and characterisation of the dopamine receptor II from the cat flea Ctenocephalides felis (CfDopRII). Insect Biochem. Mol. Biol 2006, 36, 749–758. [Google Scholar]
- Sugiura, M.; Fuke, S.; Suo, S.; Sasagawa, N.; van Tol, H.H.; Ishiura, S. Characterization of a novel D2-like dopamine receptor with a truncated splice variant and a D1-like dopamine receptor unique to invertebrates from Caenorhabditis elegans. J. Neurochem 2005, 94, 1146–1157. [Google Scholar]
- Clark, M.C.; Baro, D.J. Arthropod D2 receptors positively couple with cAMP through the Gi/o protein family. Comp. Biochem. Physiol. B 2007, 146, 9–19. [Google Scholar]
- Minneman, K.P. Splice variants of G protein-coupled receptors. Mol. Interv 2001, 1, 108–116. [Google Scholar]
- Hauser, F.; Cazzamali, G.; Williamson, M.; Blenau, W.; Grimmelikhuijzen, C.J. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog. Neurobiol 2006, 80, 1–19. [Google Scholar]
- Mitsumasu, K.; Ohta, H.; Tsuchihara, K.; Asaoka, K.; Ozoe, Y.; Niimi, T.; Yamashita, O.; Yaginuma, T. Molecular cloning and characterization of cDNAs encoding dopamine receptor-1 and -2 from brain-suboesophageal ganglion of the silkworm Bombyx mori. Insect Mol. Biol 2008, 17, 185–195. [Google Scholar]
- Clark, M.C.; Baro, D.J. Molecular cloning and characterization of crustacean type-one dopamine receptors: D1αPan and D1βPan. Comp. Biochem. Physiol. B 2006, 143, 294–301. [Google Scholar]
- Meyer, J.M.; Ejendal, K.F.; Watts, V.J.; Hill, C.A. Molecular and pharmacological characterization of two D1-like dopamine receptors in the Lyme disease vector Ixodes scapularis. Insect Biochem. Mol. Biol 2011, 41, 563–571. [Google Scholar]
- Burman, C.; Evans, P.D. Amphioxus expresses both vertebrate-type and invertebrate-type dopamine D1 receptors. Invert. Neurosci 2010, 10, 93–105. [Google Scholar]
- Defea, K. β-arrestins and heterotrimeric G-proteins: Collaborators and competitors in signal transduction. Br. J. Pharmacol 2008, 153, S298–S309. [Google Scholar]
- Ng, G.Y.; Mouillac, B.; George, S.R.; Caron, M.; Dennis, M.; Bouvier, M.; O’Dowd, B.F. Desensitization, phosphorylation and palmitoylation of the human dopamine D1 receptor. Eur. J. Pharmacol 1994, 267, 7–19. [Google Scholar]
- Ng, G.Y.; O’Dowd, B.F.; Caron, M.; Dennis, M.; Brann, M.R.; George, S.R. Phosphorylation and palmitoylation of the human D2L dopamine receptor in Sf9 cells. J. Neurochem 1994, 63, 1589–1595. [Google Scholar]
- Jin, H.; Xie, Z.; George, S.R.; O’Dowd, B.F. Palmitoylation occurs at cysteine 347 and cysteine 351 of the dopamine D1 receptor. Eur. J. Pharmacol 1999, 386, 305–312. [Google Scholar]
- Renner, U.; Glebov, K.; Lang, T.; Papusheva, E.; Balakrishnan, S.; Keller, B.; Richter, D.W.; Jahn, R.; Ponimaskin, E. Localization of the mouse 5-hydroxytryptamine1A receptor in lipid microdomains depends on its palmitoylation and is involved in receptor-mediated signaling. Mol. Pharmacol 2007, 72, 502–513. [Google Scholar]
- O’Dowd, B.F.; Hnatowich, M.; Caron, M.G.; Lefkowitz, R.J.; Bouvier, M. Palmitoylation of the human β2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J. Biol. Chem 1989, 264, 7564–7569. [Google Scholar]
- Papoucheva, E.; Dumuis, A.; Sebben, M.; Richter, D.W.; Ponimaskin, E.G. The 5-hydroxytryptamine(1A) receptor is stably palmitoylated, and acylation is critical for communication of receptor with Gi protein. J. Biol. Chem 2004, 279, 3280–32891. [Google Scholar]
- Ponimaskin, E.; Dumuis, A.; Gaven, F.; Barthet, G.; Heine, M.; Glebov, K.; Richter, D.W.; Oppermann, M. Palmitoylation of the 5-hydroxytryptamine4a receptor regulates receptor phosphorylation, desensitization, and β-arrestin-mediated endocytosis. Mol. Pharmacol 2005, 67, 1434–1443. [Google Scholar]
- Kobe, F.; Renner, U.; Woehler, A.; Wlodarczyk, J.; Papusheva, E.; Bao, G.; Zeug, A.; Richter, D.W.; Neher, E.; Ponimaskin, E. Stimulation- and palmitoylation-dependent changes in oligomeric conformation of serotonin 5-HT1A receptors. Biochim. Biophys. Acta 2008, 1783, 1503–1516. [Google Scholar]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta 2007, 1768, 794–807. [Google Scholar]
- Barak, L.S.; Tiberi, M.; Freedman, N.J.; Kwatra, M.M.; Lefkowitz, R.J.; Caron, M.G. A highly conserved tyrosine residue in G protein-coupled receptors is required for agonist-mediated β2-adrenergic receptor sequestration. J. Biol. Chem 1994, 269, 2790–2795. [Google Scholar]
- Fritze, O.; Filipek, S.; Kuksa, V.; Palczewski, K.; Hofmann, K.P.; Ernst, O.P. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc. Natl. Acad. Sci. USA 2003, 100, 2290–2295. [Google Scholar]
- Johnson, M.S.; Robertson, D.N.; Holland, P.J.; Lutz, E.M.; Mitchell, R. Role of the conserved NPxxY motif of the 5-HT2A receptor in determining selective interaction with isoforms of ADP-ribosylation factor (ARF). Cell Signal 2006, 18, 1793–1800. [Google Scholar]
- Borroto-Escuela, D.O.; Romero-Fernandez, W.; García-Negredo, G.; Correia, P.A.; Garriga, P.; Fuxe, K.; Ciruela, F. Dissecting the conserved NPxxY motif of the M3 muscarinic acetylcholine receptor: Critical role of Asp-7.49 for receptor signaling and multiprotein complex formation. Cell. Physiol. Biochem 2011, 28, 1009–1022. [Google Scholar]
- Ohta, H.; Tsuchihara, K.; Mitsumasu, K.; Yaginuma, T.; Ozoe, Y.; Asaoka, K. Comparative pharmacology of two D1-like dopamine receptors cloned from the silkworm Bombyx mori. Insect Biochem. Mol. Biol 2009, 39, 342–347. [Google Scholar]
- Reale, V.; Hannan, F.; Hall, L.M.; Evans, P.D. Agonist-specific coupling of a cloned Drosophila melanogaster D1-like dopamine receptor to multiple second messenger pathways by synthetic agonists. J. Neurosci 1997, 17, 6545–6553. [Google Scholar]
- Schmauss, C.; Haroutunian, V.; Davis, K.L.; Davidson, M. Selective loss of dopamine D3-type receptor mRNA expression in parietal and motor cortices of patients with chronic schizophrenia. Proc. Natl. Acad. Sci. USA 1993, 90, 8942–8946. [Google Scholar]
- Schmauss, C. Enhanced cleavage of an atypical intron of dopamine D3-receptor pre-mRNA in chronic schizophrenia. J. Neurosci 1996, 16, 7902–7909. [Google Scholar]
- Elmhurst, J.L.; Xie, Z.; O’Dowd, B.F.; George, S.R. The splice variant D3nf reduces ligand binding to the D3 dopamine receptor: Evidence for heterooligomerization. Brain Res. Mol. Brain Res 2000, 80, 63–74. [Google Scholar]
- Karpa, K.D.; Lin, R.; Kabbani, N.; Levenson, R. The dopamine D3 receptor interacts with itself and the truncated D3 splice variant d3nf: D3-D3nf interaction causes mislocalization of D3 receptors. Mol. Pharmacol 2000, 58, 677–683. [Google Scholar]
- Lee, S.P.; Xie, Z.; Varghese, G.; Nguyen, T.; O’Dowd, B.F.; George, S.R. Oligomerization of dopamine and serotonin receptors. Neuropsychopharmacology 2000, 23, S32–S40. [Google Scholar]
- Orr, G.L.; Gole, J.W.; Notman, H.J.; Downer, R.G. Pharmacological characterisation of the dopamine-sensitive adenylate cyclase in cockroach brain: Evidence for a distinct dopamine receptor. Life Sci 1987, 41, 2705–2715. [Google Scholar]
- Blenau, W.; May, T.; Erber, J. Characterization of [3H]LSD binding to a serotonin-sensitive site in honeybee (Apis mellifera) brain. Comp. Biochem. Physiol. B 1995, 112, 377–384. [Google Scholar]
- Stengl, M.; Homberg, U. Pigment-dispersing hormone-immunoreactive neurons in the cockroach Leucophaea maderae share properties with circadian pacemaker neurons. J. Comp. Physiol. A 1994, 175, 203–213. [Google Scholar]
- Baumann, O.; Kühnel, D.; Dames, P.; Walz, B. Dopaminergic and serotonergic innervation of cockroach salivary glands: Distribution and morphology of synapses and release sites. J. Exp. Biol 2004, 207, 2565–2575. [Google Scholar]
- Lang, I.; Walz, B. Dopamine stimulates salivary duct cells in the cockroach Periplaneta americana. J. Exp. Biol 1999, 202, 729–738. [Google Scholar]
- Šimo, L.; Zitňan, D.; Park, Y. Neural control of salivary glands in ixodid ticks. J. Insect Physiol 2012, 58, 459–466. [Google Scholar]
- Šimo, L.; Koči, J.; Žitňan, D.; Park, Y. Evidence for D1 dopamine receptor activation by a paracrine signal of dopamine in tick salivary glands. PLoS One 2011, 6, e16158. [Google Scholar]
- Blenau, W.; Baumann, A. Molecular characterization of the ebony gene from the American cockroach Periplaneta americana. Arch. Insect Biochem. Physiol 2005, 59, 184–195. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
- Chen, C.; Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol 1987, 7, 2745–2752. [Google Scholar]
- Schlenstedt, J.; Balfanz, S.; Baumann, A.; Blenau, W. Am5-HT7: Molecular and pharmacological characterization of the first 5-HT receptor of the honeybee (Apis mellifera). J. Neurochem 2006, 98, 1985–1998. [Google Scholar]




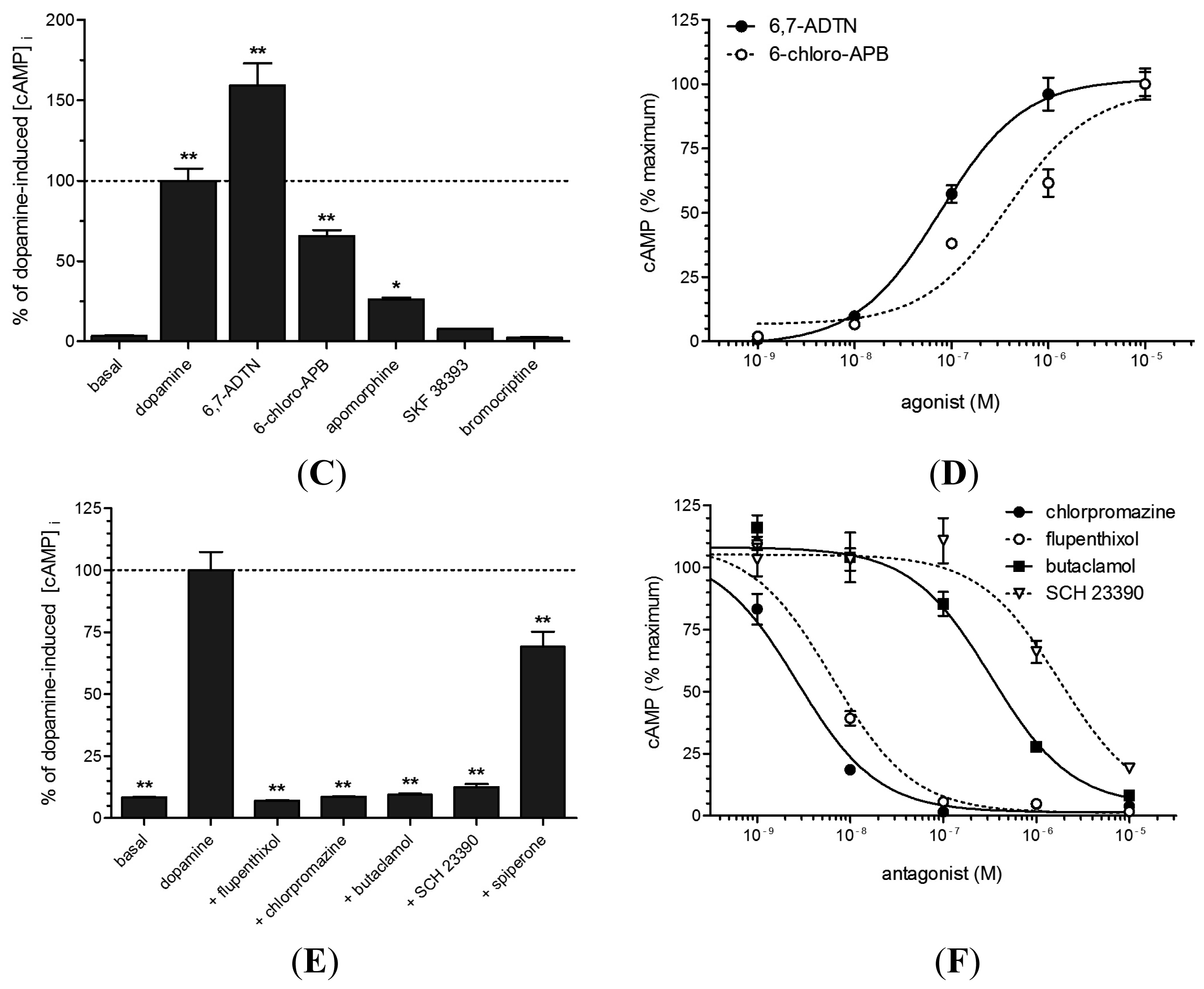
| Substance | EC50/IC50 (nM) | logEC50/log IC50 |
|---|---|---|
| agonists | ||
| dopamine | 163.3 | −6.787 |
| 6,7-ADTN | 76.4 | −7.117 |
| 6-chloro-APB | 381.9 | −6.418 |
| antagonists | ||
| chlorpromazine | 2.7 | −8.569 |
| flupentixol | 6.5 | −8.189 |
| butaclamol | 326.0 | −6.487 |
| SCH 23390 | 1786.0 | −5.748 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Troppmann, B.; Balfanz, S.; Krach, C.; Baumann, A.; Blenau, W. Characterization of an Invertebrate-Type Dopamine Receptor of the American Cockroach, Periplaneta americana. Int. J. Mol. Sci. 2014, 15, 629-653. https://doi.org/10.3390/ijms15010629
Troppmann B, Balfanz S, Krach C, Baumann A, Blenau W. Characterization of an Invertebrate-Type Dopamine Receptor of the American Cockroach, Periplaneta americana. International Journal of Molecular Sciences. 2014; 15(1):629-653. https://doi.org/10.3390/ijms15010629
Chicago/Turabian StyleTroppmann, Britta, Sabine Balfanz, Christian Krach, Arnd Baumann, and Wolfgang Blenau. 2014. "Characterization of an Invertebrate-Type Dopamine Receptor of the American Cockroach, Periplaneta americana" International Journal of Molecular Sciences 15, no. 1: 629-653. https://doi.org/10.3390/ijms15010629




