MiRNA-199a-3p Regulates C2C12 Myoblast Differentiation through IGF-1/AKT/mTOR Signal Pathway
Abstract
:1. Introduction
2. Results
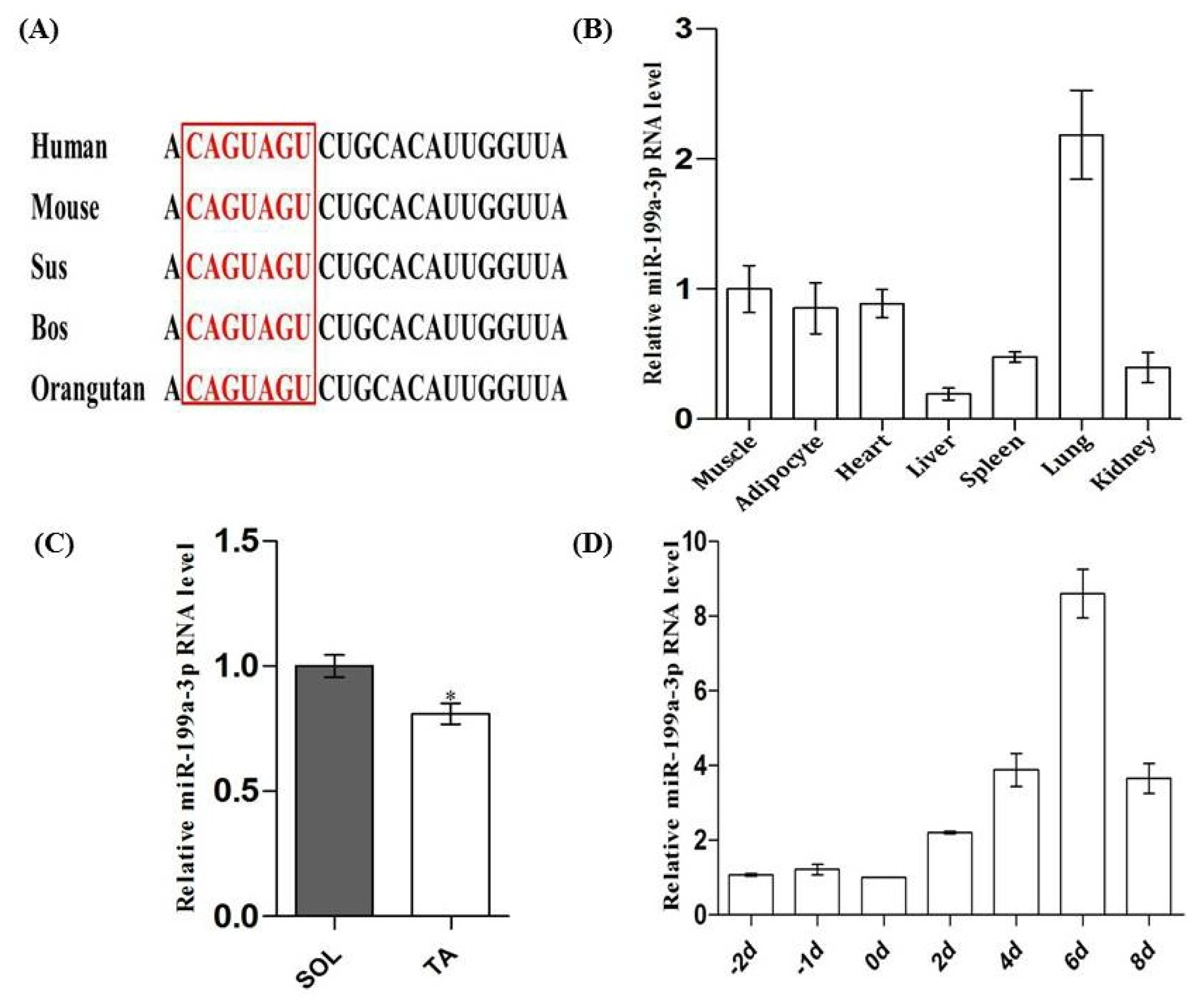
2.1. Tissue Expression Profile of MiR-199a-3p and Its Expression Pattern during C2C12 Myogenic Differentiation
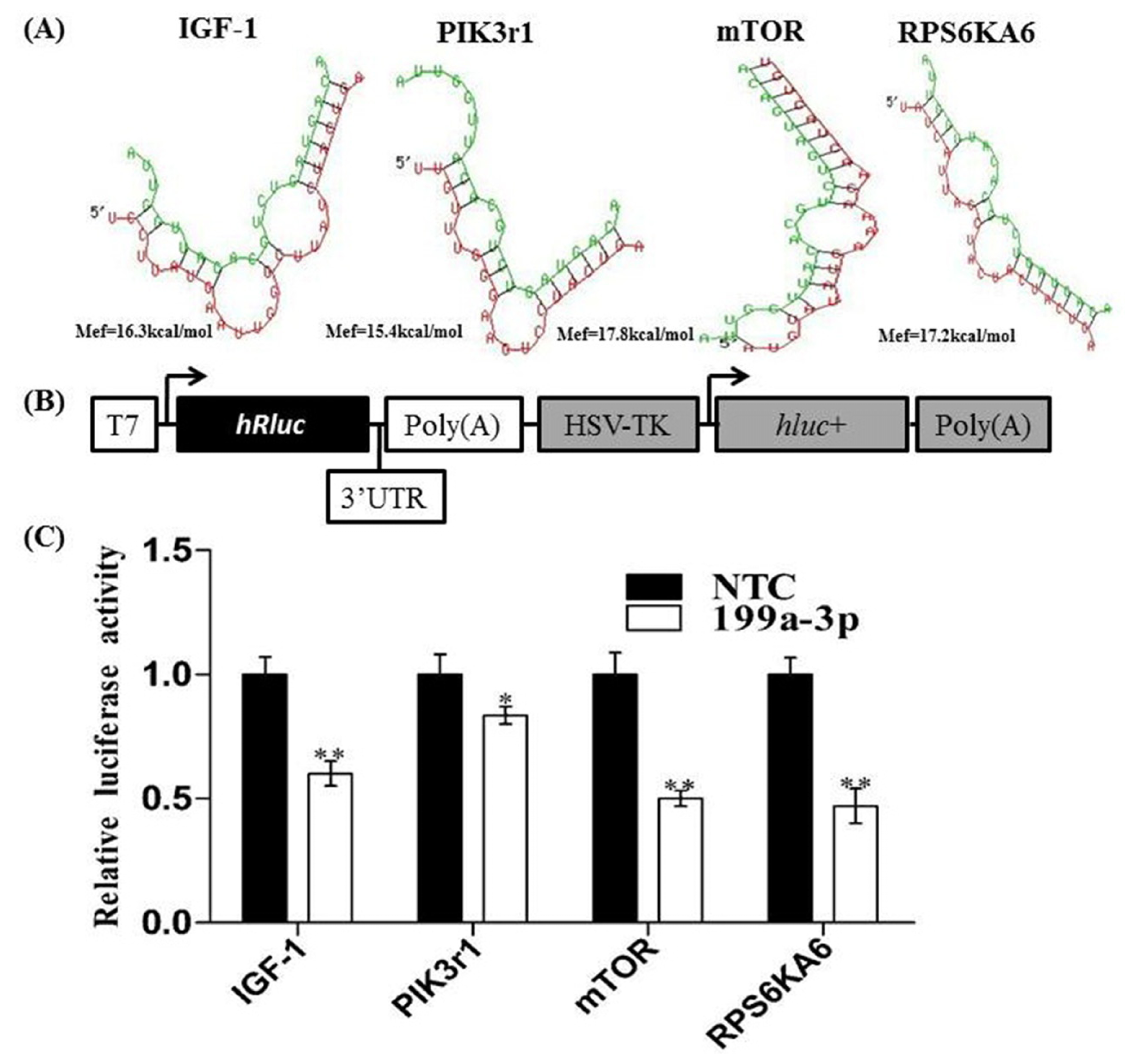
2.2. Analysis of MiR-199a-3p Target Genes
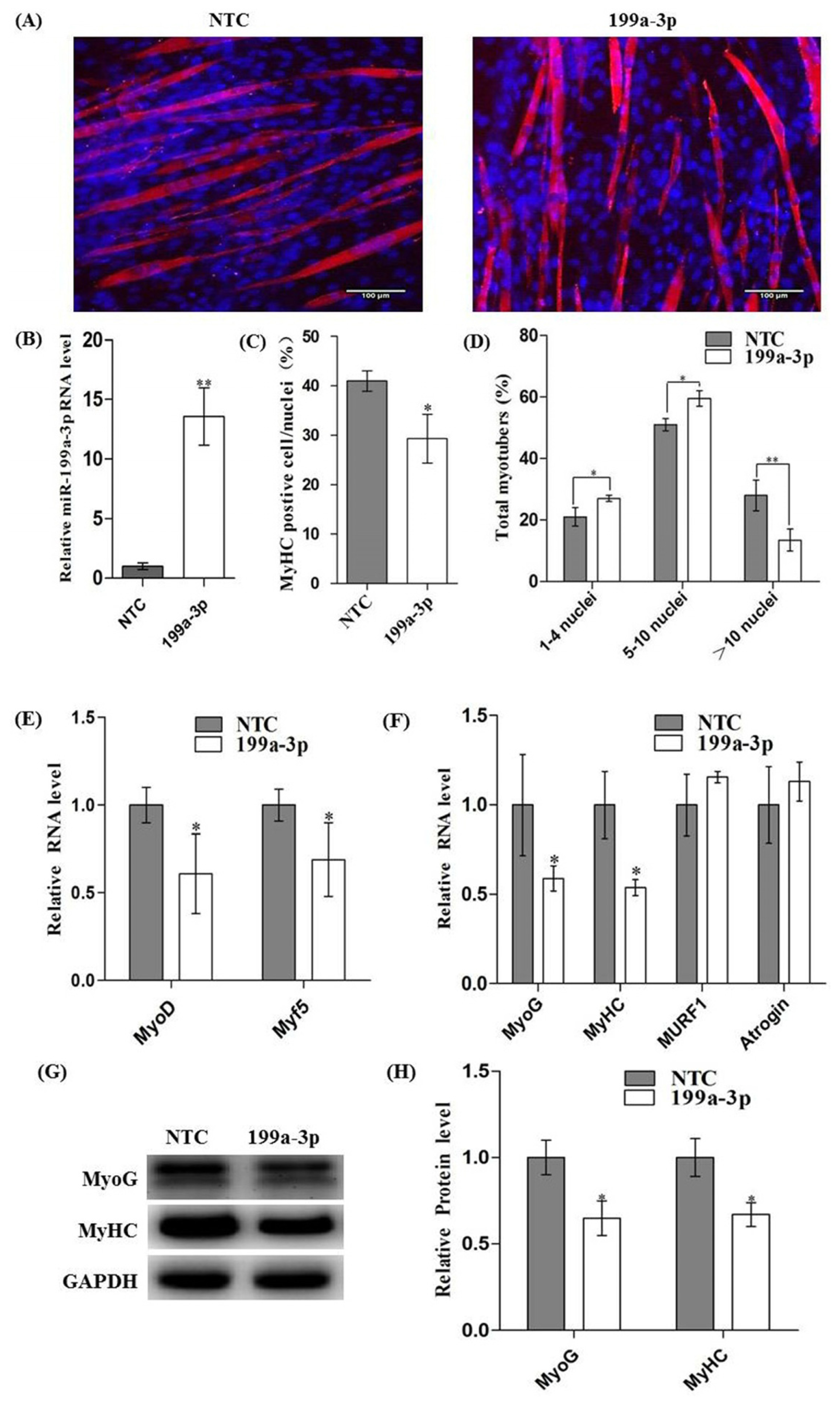
2.3. Over-Expression of MiR-199a-3p Inhibits C2C12 Myogenic Differentiation
2.4. Inhibition of MiR-199a-3p Promotes Myogenic Differentiation and Myotube Hypertrophy
2.5. MiR-199a-3p Affects IGF/AKT/mTOR Pathway in C2C12 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Transfection of MiR-199a-3p Mimic and Inhibitor
4.3. Immunocytochemical Analysis
4.4. Real-Time qPCR
4.5. Western Blot Detection
4.6. Luciferase Reporter Assay
4.7. Statistic Analysis
5. Conclusions
Supplementary Information
ijms-15-00296-s001.pdfAcknowledgments
Conflicts of Interest
References
- Kuang, S.; Kuroda, K.; le Grand, F.; Rudnicki, M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007, 129, 999–1010. [Google Scholar]
- Singh, K.; Dilworth, F.J. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J 2013, 280, 3991–4003. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Callis, T.E.; Deng, Z.; Chen, J.F.; Wang, D.Z. Muscling through the microRNA world. Exp. Biol. Med 2008, 233, 131–138. [Google Scholar]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet 2006, 38, 228–233. [Google Scholar]
- Sweetman, D.; Goljanek, K.; Rathjen, T.; Oustanina, S.; Braun, T.; Dalmay, T.; Münsterberg, A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol 2008, 321, 491–499. [Google Scholar] [Green Version]
- Dey, B.K.; Gagan, J.; Yan, Z.; Dutta, A. MiR-26a is required for skeletal muscle differentiation and regeneration in mice. Gene. Dev 2012, 26, 2180–2191. [Google Scholar]
- Crist, C.G.; Montarras, D.; Pallafacchina, G.; Rocancourt, D.; Cumano, A.; Conway, S.J.; Buckingham, M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci.USA 2009, 106, 13383–13387. [Google Scholar]
- Wei, W.; He, H.B.; Zhang, W.Y.; Zhang, H.X.; Bai, J.B.; Liu, H.Z.; Cao, J.H.; Chang, K.C.; Li, X.Y.; Zhao, S.H. MiR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis 2013, 4, e668. [Google Scholar]
- Ge, Y.; Sun, Y.; Chen, J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol 2011, 192, 69–81. [Google Scholar]
- Seok, H.Y.; Tatsuguchi, M.; Callis, T.E.; He, A.; Pu, W.T.; Wang, D.Z. MiR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J. Biol. Chem 2011, 286, 35339–35346. [Google Scholar]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, R.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol 2006, 8, 278–284. [Google Scholar]
- Feng, Y.; Cao, J.H.; Li, X.Y.; Zhao, S.H. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochem. Funct 2011, 29, 378–383. [Google Scholar]
- Juan, A.H.; Kumar, R.M.; Marx, J.G.; Young, R.A.; Sartorelli, V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 2009, 36, 61–74. [Google Scholar]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar]
- Kierdaszuk, B.; Berdynski, M.; Karolczak, J.; Redowicz, M.J.; Zekanowski, C.; Kaminska, A.M. A novel mutation in the DNM2 gene impairs dynamin 2 localization in skeletal muscle of a patient with late onset centronuclear myopathy. Neuromuscul. Disord 2013, 23, 219–228. [Google Scholar]
- Watanabe, T.; Sato, T.; Amano, T.; Kawamura, Y.; Kawamura, N.; Kawaguchi, H.; Yamashita, N.; Kurihara, H.; Nakaoka, T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn 2008, 23, 3738–3748. [Google Scholar]
- Tian, W.; Dong, X.; Liu, X.; Wang, G.; Dong, Z.; Shen, W.; Zheng, G.; Lu, J.; Chen, J.; Wang, Y.; et al. High-throughput functional microRNAs profiling by recombinant AAV-based microRNA sensor arrays. PLoS One 2012, 7, e29551. [Google Scholar]
- Roberts, T.C.; Blomberg, K.E.; McClorey, G.; Andaloussi, S.; Godfrey, C.; Betts, C.; Coursindel, T.; Gait, M.J.; Smith, C.I.; Wood, M.J. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol. Ther 2012, 1, e39. [Google Scholar]
- Mungunsukh, O.; DayR, M. Transforming growth factor-β1 selectively inhibits hepatocyte growth factor expression via a micro-RNA-199-dependent posttranscriptional mechanism. Mol. Biol. Cell 2013, 24, 2088–2097. [Google Scholar]
- Philippou, A.; Maridaki, M.; Halapas, A.; Koutsilieris, M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo 2007, 21, 45–54. [Google Scholar]
- Jones, N.C.; Fedorov, Y.V.; Rosenthal, R.S.; Olwin, B.B. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J. Cell. Physiol 2001, 118, 104–115. [Google Scholar]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol 2001, 3, 1014–1019. [Google Scholar]
- Elia, L.; Contu, R.; Quintavalle, M.; Varrone, F.; Chimenti, C.; Russo, M.A.; Cimino, V.; de Marinis, L.; Frustaci, A.; Catalucci, D.; et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 2009, 120, 2377–2385. [Google Scholar]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Sci 2013, 126, 2678–2691. [Google Scholar]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci.USA 2010, 107, 4218–4223. [Google Scholar]
- Wu, J.H.; Gao, Y.; Ren, A.J.; Zhao, S.H.; Zhong, M.; Peng, Y.J.; Shen, W.; Jing, M.; Liu, L. Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res 2012, 47, 195–201. [Google Scholar]
- Santhakumar, D.; Forster, T.; Laqtom, N.N.; Fragkoudis, R.; Dickinson, P.; Abreu-Goodger, C.; Manakov, A.; Choudhury, N.R.; Griffiths, S.J.; Vermeulen, A.; et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 13830–13835. [Google Scholar] [Green Version]
- Bouzakri, K.; Zachrisson, A.; Al-Khalili, L.; Zhang, B.B.; Koistinen, H.A.; Krook, A.; Zierath, J.R. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab 2006, 4, 89–96. [Google Scholar]
- Akhtar, N.; Haqqi, T.M. MicroRNA-199a* regulates the expression of cyclooxygenase-2 in human chondrocytes. Ann. Rheum. Dis 2012, 71, 1073–1080. [Google Scholar]


© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jia, L.; Li, Y.-F.; Wu, G.-F.; Song, Z.-Y.; Lu, H.-Z.; Song, C.-C.; Zhang, Q.-L.; Zhu, J.-Y.; Yang, G.-S.; Shi, X.-E. MiRNA-199a-3p Regulates C2C12 Myoblast Differentiation through IGF-1/AKT/mTOR Signal Pathway. Int. J. Mol. Sci. 2014, 15, 296-308. https://doi.org/10.3390/ijms15010296
Jia L, Li Y-F, Wu G-F, Song Z-Y, Lu H-Z, Song C-C, Zhang Q-L, Zhu J-Y, Yang G-S, Shi X-E. MiRNA-199a-3p Regulates C2C12 Myoblast Differentiation through IGF-1/AKT/mTOR Signal Pathway. International Journal of Molecular Sciences. 2014; 15(1):296-308. https://doi.org/10.3390/ijms15010296
Chicago/Turabian StyleJia, Long, Yue-Feng Li, Guo-Fang Wu, Zi-Yi Song, Hong-Zhao Lu, Cheng-Chuang Song, Qiang-Ling Zhang, Jia-Yu Zhu, Gong-She Yang, and Xin-E Shi. 2014. "MiRNA-199a-3p Regulates C2C12 Myoblast Differentiation through IGF-1/AKT/mTOR Signal Pathway" International Journal of Molecular Sciences 15, no. 1: 296-308. https://doi.org/10.3390/ijms15010296




