DNA Repair Gene XRCC4 Codon 247 Polymorphism Modified Diffusely Infiltrating Astrocytoma Risk and Prognosis
Abstract
:1. Introduction
2. Results
2.1. Demographic and Clinic Characteristics of the Subjects
2.2. XRCC4 Codon 247 Polymorphism Increased DIA Risk
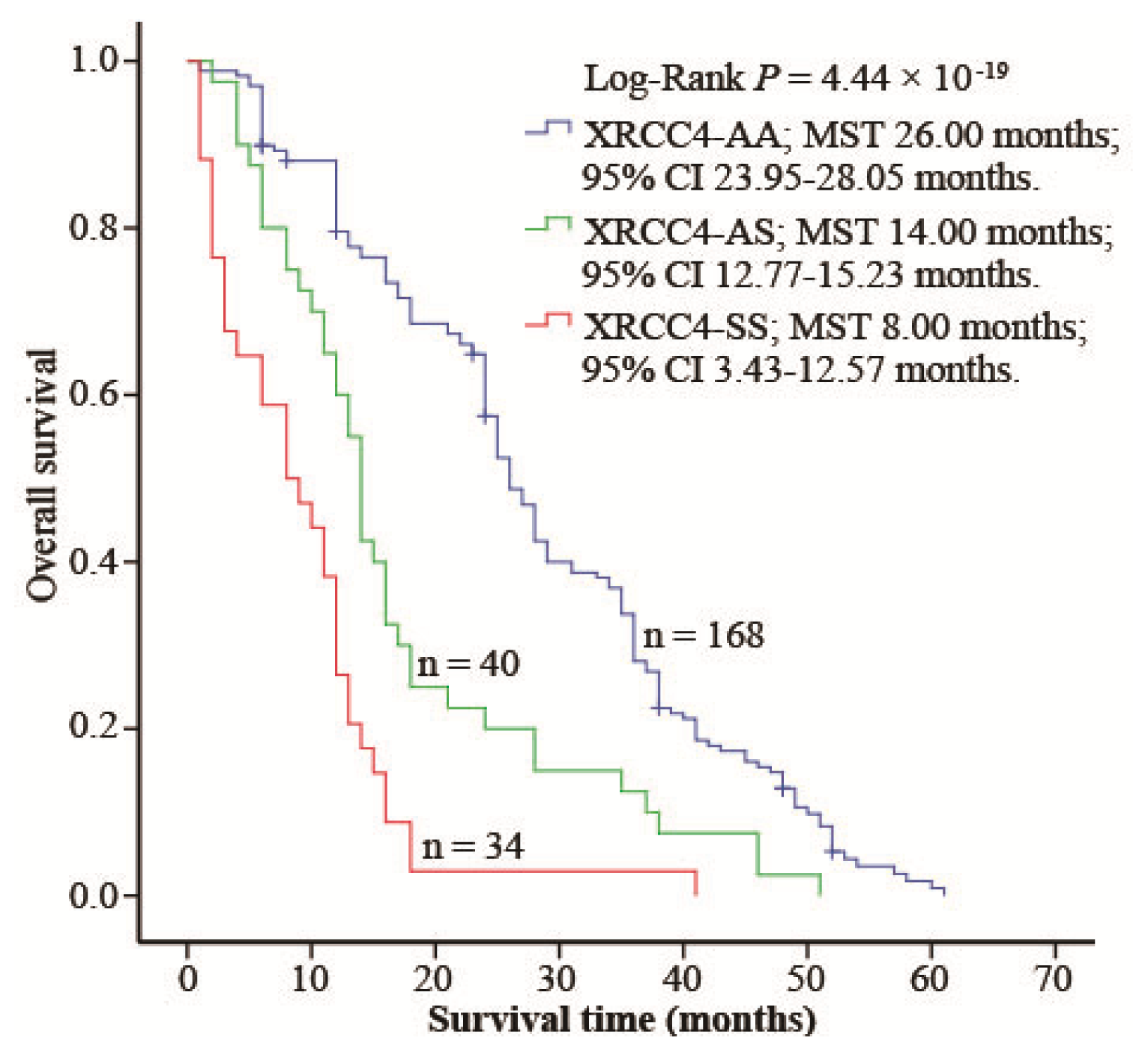
2.4. XRCC4 Codon 247 Polymorphism Modified DIA Prognosis
3. Discussion
4. Experimental Section
4.1. Study Population
4.2. DNA Extraction
4.3. Genotyping
4.4. DIA Patients Follow-up
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ohgaki, H.; Kleihues, P. Epidemiology and etiology of gliomas. Acta Neuropathol 2005, 109, 93–108. [Google Scholar]
- Tzeng, S.Y.; Green, J.J. Therapeutic nanomedicine for brain cancer. Ther. Deliv 2013, 4, 687–704. [Google Scholar]
- Ohgaki, H.; Kleihues, P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci 2009, 100, 2235–2241. [Google Scholar]
- Ohgaki, H.; Kleihues, P. The definition of primary and secondary glioblastoma. Clin. Cancer Res 2013, 19, 764–772. [Google Scholar]
- Weller, M.; Pfister, S.M.; Wick, W.; Hegi, M.E.; Reifenberger, G.; Stupp, R. Molecular neuro-oncology in clinical practice: A new horizon. Lancet Oncol 2013, 14, e370–e379. [Google Scholar]
- Wang, L.E.; Bondy, M.L.; Shen, H.; El-Zein, R.; Aldape, K.; Cao, Y.; Pudavalli, V.; Levin, V.A.; Yung, W.K.; Wei, Q. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res 2004, 64, 5560–5563. [Google Scholar]
- Premkumar, D.R.; Jane, E.P.; Agostino, N.R.; DiDomenico, J.D.; Pollack, I.F. Bortezomib-induced sensitization of malignant human glioma cells to vorinostat-induced apoptosis depends on reactive oxygen species production, mitochondrial dysfunction, Noxa upregulation, Mcl-1 cleavage, and DNA damage. Mol. Carcinog 2013, 52, 118–133. [Google Scholar]
- Spyropoulou, A.; Piperi, C.; Adamopoulos, C.; Papavassiliou, A.G. Deregulated chromatin remodeling in the pathobiology of brain tumors. Neuromol. Med 2013, 15, 1–24. [Google Scholar]
- Wang, C.; Lees-Miller, S.P. Detection and repair of ionizing radiation-induced DNA double strand breaks: New developments in nonhomologous end joining. Int. J. Radiat. Oncol. Biol. Phys 2013, 86, 440–449. [Google Scholar]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res 2013, 2, 130–143. [Google Scholar]
- Zhao, B.; Ye, J.; Li, B.; Ma, Q.; Su, G.; Han, R. DNA repair gene XRCC3 Thr241Met polymorphism and glioma risk: A meta-analysis. Int. J. Clin. Exp. Med 2013, 6, 438–443. [Google Scholar]
- Custodio, A.C.; Almeida, L.O.; Pinto, G.R.; Santos, M.J.; Almeida, J.R.; Clara, C.A.; Rey, J.A.; Casartelli, C. Variation in DNA repair gene XRCC3 affects susceptibility to astrocytomas and glioblastomas. Genet. Mol. Res 2012, 11, 332–339. [Google Scholar] [Green Version]
- Berntsson, S.G.; Wibom, C.; Sjostrom, S.; Henriksson, R.; Brannstrom, T.; Broholm, H.; Johansson, C.; Fleming, S.J.; McKinney, P.A.; Bethke, L.; et al. Analysis of DNA repair gene polymorphisms and survival in low-grade and anaplastic gliomas. J. Neurooncol 2011, 105, 531–538. [Google Scholar]
- Wu, P.Y.; Frit, P.; Meesala, S.; Dauvillier, S.; Modesti, M.; Andres, S.N.; Huang, Y.; Sekiguchi, J.; Calsou, P.; Salles, B.; et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol. Cell. Biol 2009, 29, 3163–3172. [Google Scholar]
- Wu, C.N.; Liang, S.Y.; Tsai, C.W.; Bau, D.T. The role of XRCC4 in carcinogenesis and anticancer drug discovery. Recent Patents Anti-Cancer Drug Discovery 2008, 3, 209–219. [Google Scholar]
- Pardo, B.; Gomez-Gonzalez, B.; Aguilera, A. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell. Mol. Life Sci 2009, 66, 1039–1056. [Google Scholar]
- Shao, N.; Jiang, W.Y.; Qiao, D.; Zhang, S.G.; Wu, Y.; Zhang, X.X.; Hua, L.X.; Ding, Y.; Feng, N.H. An updated meta-analysis of XRCC4 polymorphisms and cancer risk based on 31 case-control studies. Cancer Biomark 2012, 12, 37–47. [Google Scholar]
- Long, X.D.; Zhao, D.; Wang, C.; Huang, X.Y.; Yao, J.G.; Ma, Y.; Wei, Z.H.; Liu, M.; Zeng, L.X.; Mo, X.Q.; et al. Genetic polymorphisms in DNA repair genes XRCC4 and XRCC5 and aflatoxin B1-related hepatocellular carcinoma. Epidemiology 2013, 24, 671–681. [Google Scholar]
- Long, X.D.; Yao, J.G.; Zeng, Z.; Ma, Y.; Huang, X.Y.; Wei, Z.H.; Liu, M.; Zhang, J.J.; Xue, F.; Zhai, B.; et al. Polymorphisms in the coding region of X-ray repair complementing group 4 and aflatoxin B1-related hepatocellular carcinoma. Hepatology 2013, 58, 171–181. [Google Scholar]
- Tseng, H.C.; Tsai, M.H.; Chiu, C.F.; Wang, C.H.; Chang, N.W.; Huang, C.Y.; Tsai, C.W.; Liang, S.Y.; Wang, C.L.; Bau, D.T. Association of XRCC4 codon 247 polymorphism with oral cancer susceptibility in Taiwan. Anticancer Res 2008, 28, 1687–1691. [Google Scholar]
- Wibom, C.; Sjostrom, S.; Henriksson, R.; Brannstrom, T.; Broholm, H.; Ryden, P.; Johansen, C.; Collatz-Laier, H.; Hepworth, S.; McKinney, P.A.; et al. DNA-repair gene variants are associated with glioblastoma survival. Acta Oncol 2012, 51, 325–332. [Google Scholar]
- Mahaney, B.L.; Hammel, M.; Meek, K.; Tainer, J.A.; Lees-Miller, S.P. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol 2013, 91, 31–41. [Google Scholar]
- van Heemst, D.; Brugmans, L.; Verkaik, N.S.; van Gent, D.C. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair 2004, 3, 43–50. [Google Scholar]
- Gao, Y.; Sun, Y.; Frank, K.M.; Dikkes, P.; Fujiwara, Y.; Seidl, K.J.; Sekiguchi, J.M.; Rathbun, G.A.; Swat, W.; Wang, J.; et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 1998, 95, 891–902. [Google Scholar]
- Gao, Y.; Ferguson, D.O.; Xie, W.; Manis, J.P.; Sekiguchi, J.; Frank, K.M.; Chaudhuri, J.; Horner, J.; DePinho, R.A.; Alt, F.W. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 2000, 404, 897–900. [Google Scholar]
- Long, X.D.; Ma, Y.; Huang, Y.Z.; Yi, Y.; Liang, Q.X.; Ma, A.M.; Zeng, L.P.; Fu, G.H. Genetic polymorphisms in DNA repair genes XPC, XPD, and XRCC4, and susceptibility to Helicobacter pylori infection-related gastric antrum adenocarcinoma in Guangxi population, China. Mol. Carcinog 2010, 49, 611–618. [Google Scholar]
- Chiu, C.F.; Wang, C.H.; Wang, C.L.; Lin, C.C.; Hsu, N.Y.; Weng, J.R.; Bau, D.T. A novel single nucleotide polymorphism in XRCC4 gene is associated with gastric cancer susceptibility in Taiwan. Ann. Surg. Oncol 2008, 15, 514–518. [Google Scholar]
- Figueroa, J.D.; Malats, N.; Rothman, N.; Real, F.X.; Silverman, D.; Kogevinas, M.; Chanock, S.; Yeager, M.; Welch, R.; Dosemeci, M.; et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis 2007, 28, 1788–1793. [Google Scholar]
- Fan, X.J.; Ren, P.L.; Lu, Z.J.; Zhao, S.; Yang, X.L.; Liu, J. The study of esophageal cancer risk associated with polymorphisms of DNA damage repair genes XRCC4 and RAD51. Sichuan Da Xue Xue Bao Yi Xue Ban 2013, 44, 568–572. [Google Scholar]
| Characteristics | Controls (n = 358) | Cases (n = 242) | χ2 | p | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Age (years) | 0.317 | 1.000 | ||||
| ≤35 | 19 | 5.3 | 13 | 5.4 | ||
| 36–40 | 23 | 6.4 | 16 | 6.6 | ||
| 41–45 | 33 | 9.2 | 22 | 9.1 | ||
| 46–50 | 76 | 21.2 | 53 | 21.9 | ||
| 51–55 | 102 | 28.5 | 68 | 28.1 | ||
| 56–60 | 45 | 12.6 | 30 | 12.4 | ||
| 61–65 | 34 | 9.5 | 24 | 9.9 | ||
| ≥66 | 26 | 7.3 | 15 | 6.2 | ||
| Sex | 0.318 | 0.573 | ||||
| Male | 230 | 64.2 | 150 | 62.0 | ||
| Female | 128 | 35.8 | 92 | 38.0 | ||
| Race | 0.026 | 0.872 | ||||
| Han | 184 | 51.4 | 126 | 52.1 | ||
| Minority | 174 | 48.6 | 116 | 47.9 | ||
| XRCC4 | Controls | Cases | OR (95%CI) | p | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Genotype | AA b | 298 | 83.2 | 168 | 69.4 | 1 | |
| AS b | 39 | 10.9 | 40 | 16.5 | 1.82(1.13–2.94) a | 1.46 × 10−3 | |
| SS b | 21 | 5.9 | 34 | 14.0 | 2.89(1.62–5.15) a | 3.22 × 10−4 | |
| Allele | Ala c | 635 | 88.7 | 376 | 77.7 | 1 | |
| Ser d | 81 | 11.3 | 108 | 22.3 | 2.25(1.64–3.09) | 4.43 × 10−7 | |
| Variable | Genotype | Control | Case | OR (95%CI) a | p | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Race b | XRCC4 | ||||||
| Han | AA | 152 | 82.6 | 87 | 69.0 | 1 | |
| AS | 21 | 11.4 | 21 | 16.7 | 1.73(0.89–3.34) | 0.11 | |
| SS | 11 | 5.8 | 18 | 14.2 | 3.07(1.35–7.00) | 7.68 × 10−3 | |
| AS/SS | 32 | 17.2 | 39 | 30.9 | 2.16(1.25–3.71) | 5.65 × 10−3 | |
| Zhuang | AA | 146 | 83.9 | 81 | 69.6 | 1 | |
| AS | 18 | 10.2 | 19 | 16.4 | 1.87(0.93–3.76) | 0.08 | |
| SS | 11 | 6.1 | 16 | 14.1 | 2.61(1.16–5.92) | 0.02 | |
| AS/SS | 28 | 16.3 | 35 | 30.5 | 2.15(1.22–3.78) | 7.89 × 10−3 | |
| Gender c | XRCC4 | ||||||
| Male | AA | 193 | 83.9 | 104 | 69.3 | 1 | |
| AS | 24 | 10.4 | 27 | 18.0 | 2.10(1.15–3.84) | 1.56 × 10−2 | |
| SS | 13 | 5.8 | 19 | 12.7 | 2.81(1.33–5.96) | 6.95 × 10−3 | |
| AS/SS | 37 | 16.2 | 46 | 30.7 | 2.35(1.43–3.86) | 7.82 × 10−4 | |
| Female | AA | 105 | 82.0 | 64 | 69.3 | 1 | |
| AS | 15 | 11.5 | 13 | 14.1 | 1.40(0.62–3.15) | 0.42 | |
| SS | 8 | 6.2 | 15 | 16.5 | 2.86(1.14–7.19) | 2.50 × 10−2 | |
| AS/SS | 23 | 17.8 | 28 | 30.6 | 1.92(1.01–3.64) | 4.60 × 10−2 | |
| Age d | XRCC4 | ||||||
| ≤50 | AA | 128 | 84.8 | 75 | 72.1 | 1 | |
| AS | 17 | 11.3 | 18 | 17.3 | 1.83(0.89–3.78) | 0.10 | |
| SS | 6 | 3.8 | 11 | 10.6 | 3.19(1.13–9.02) | 2.85 × 10−2 | |
| AS/SS | 151 | 99.9 | 29 | 27.9 | 2.19(1.18–4.06) | 1.34 × 10−2 | |
| >50 | AA | 170 | 82.1 | 93 | 67.2 | 1 | |
| AS | 22 | 10.5 | 22 | 15.9 | 1.86(0.98–3.56) | 0.06 | |
| SS | 15 | 7.3 | 23 | 16.8 | 2.76(1.37–5.56) | 4.56 × 10−3 | |
| AS/SS | 37 | 17.9 | 45 | 32.7 | 2.23(1.35–3.70) | 1.88 × 10−3 | |
| II | III | IV | ||||
|---|---|---|---|---|---|---|
| XRCC4 | n | % | n | % | n | % |
| AA | 42 | 89.4 | 41 | 77.4 | 85 | 59.7 |
| AS | 4 | 8.5 | 7 | 13.2 | 29 | 20.4 |
| SS | 1 | 2.1 | 5 | 9.4 | 28 | 19.9 |
| AS/SS | 5 | 10.6 | 12 | 22.6 | 57 | 40.3 |
| Variable | HR(95%CI) | p |
|---|---|---|
| XRCC4 | ||
| AA | 1 | |
| AS | 2.26(1.57–3.24) | 9.75 × 10−6 |
| SS | 5.36(3.51–8.17) | 6.42 × 10−15 |
| Grade | ||
| II | 1 | |
| III | 4.27(2.53–7.20) | 5.32 × 10−8 |
| IV | 9.27(5.54–15.51) | 2.31 × 10−17 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lin, Z.-H.; Chen, J.-C.; Wang, Y.-S.; Huang, T.-J.; Wang, J.; Long, X.-D. DNA Repair Gene XRCC4 Codon 247 Polymorphism Modified Diffusely Infiltrating Astrocytoma Risk and Prognosis. Int. J. Mol. Sci. 2014, 15, 250-260. https://doi.org/10.3390/ijms15010250
Lin Z-H, Chen J-C, Wang Y-S, Huang T-J, Wang J, Long X-D. DNA Repair Gene XRCC4 Codon 247 Polymorphism Modified Diffusely Infiltrating Astrocytoma Risk and Prognosis. International Journal of Molecular Sciences. 2014; 15(1):250-260. https://doi.org/10.3390/ijms15010250
Chicago/Turabian StyleLin, Zhong-Hui, Jin-Chun Chen, Yun-Sun Wang, Teng-Jiao Huang, Jin Wang, and Xi-Dai Long. 2014. "DNA Repair Gene XRCC4 Codon 247 Polymorphism Modified Diffusely Infiltrating Astrocytoma Risk and Prognosis" International Journal of Molecular Sciences 15, no. 1: 250-260. https://doi.org/10.3390/ijms15010250




