A One-Step Homogeneous Sandwich Immunosensor for Salmonella Detection Based on Magnetic Nanoparticles (MNPs) and Quantum Dots (QDs)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
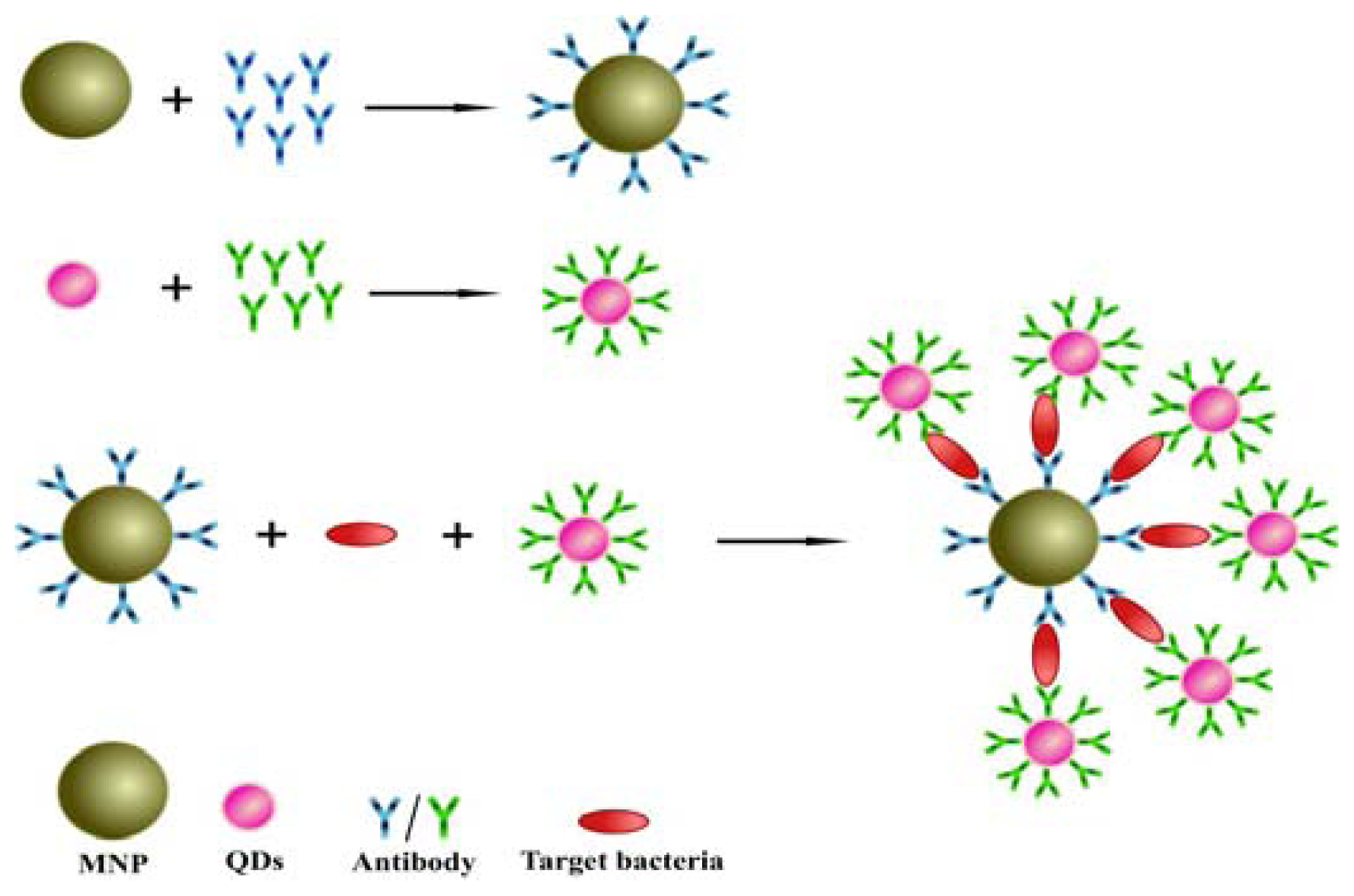
2.2. Design of the Sandwich Immunoassay for Salmonella Detection
2.3 Functionalization of MNPs with Antibody
2.3.1. Synthesis of QDs-Ab Conjugates
2.3.2. Salmonella Sample Analysis
3. Results and Discussion
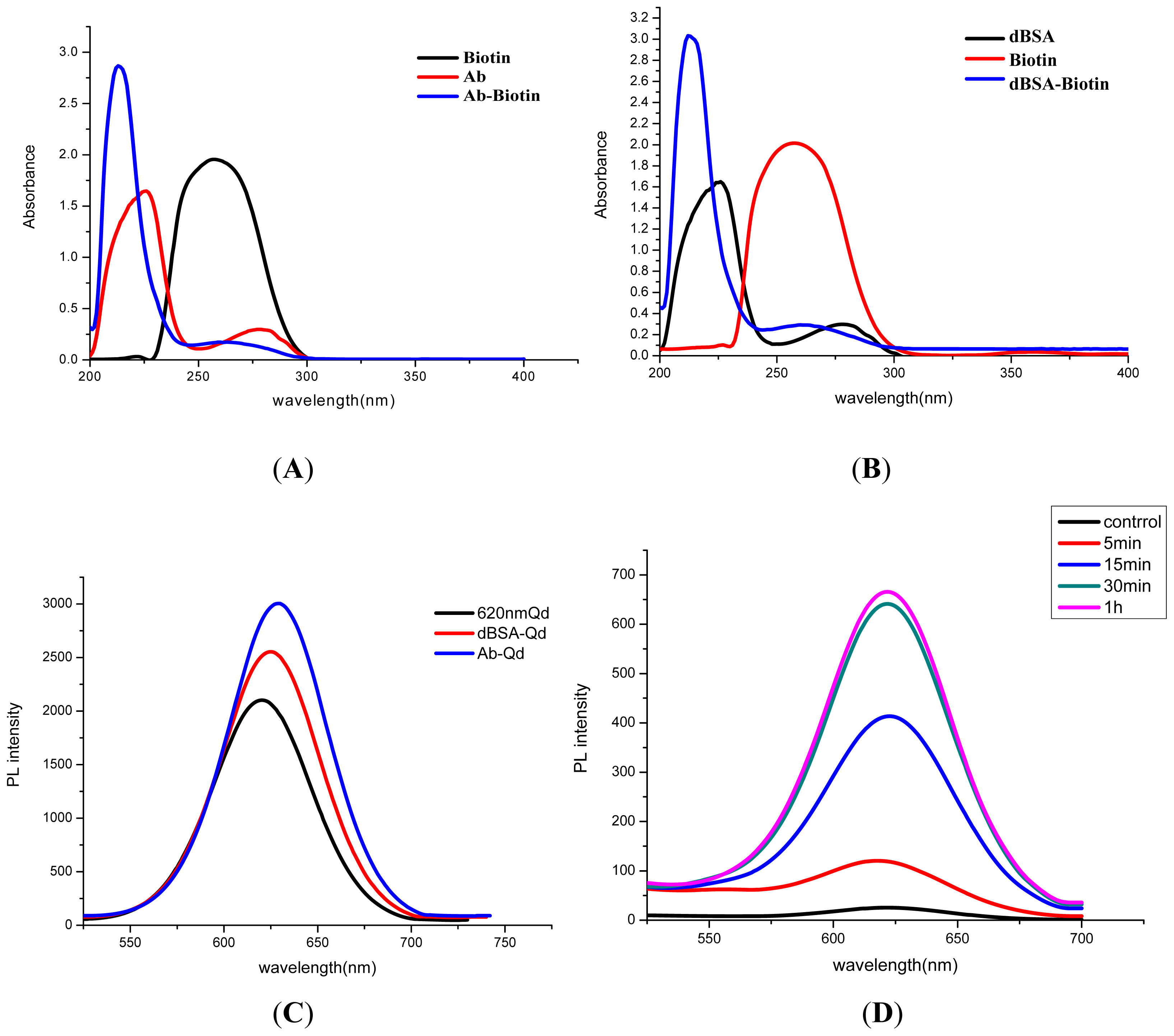
3.1. Characterization of QD-Ab
3.2. Optimization of the Immunoassay System
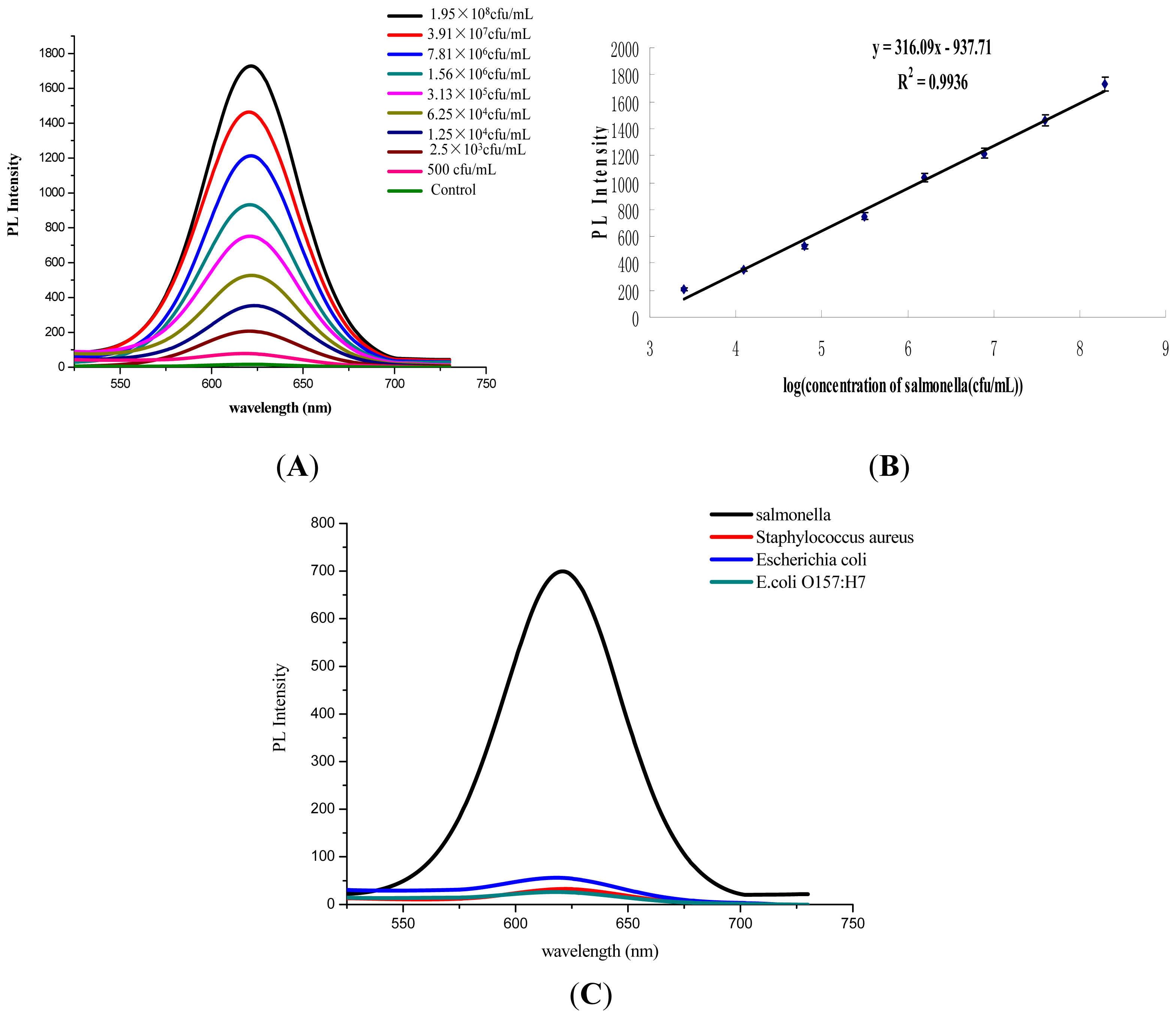
3.3. Performance Assessment of Salmonella Detection
4. Conclusions
Acknowledgments
References
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases-The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol 2010, 139, S3–S15. [Google Scholar]
- Wong, T.L.; MacDiarmid, S.; Cook, R. Salmonella, Escherichia coli O157:H7 and E. coli biotype 1 in a pilot survey of imported and New Zealand pig meats. Food Microbiol 2009, 26, 177–182. [Google Scholar]
- Switt, A.I.M.; Soyer, Y.; Warnick, L.D.; Wiedmann, M. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:-. Foodborne Pathog. Dis 2009, 6, 407–415. [Google Scholar]
- Abdoel, T.H.; Pastoor, R.; Smits, H.L.; Hatta, M. Laboratory evaluation of a simple and rapid latex agglutination assay for the serodiagnosis of typhoid fever. Trans. R. Soc. Trop. Med. Hyg 2007, 101, 1032–1038. [Google Scholar]
- Fraser, A.; Paul, M.; Goldberg, E.; Acosta, C.J.; Leibovici, L. Typhoid fever vaccines: Systematic review and meta-analysis of randomised controlled trials. Vaccine 2007, 25, 7848–7857. [Google Scholar]
- Tauxe, R.V. Salmonella: A postmodern pathogen. J. Food Prot 1991, 54, 563–568. [Google Scholar]
- Ghenghesh, K.S.; Franka, E.; Tawil, K.; Wasfy, M.O.; Ahmed, S.F.; Rubino, S.; Klena, J.D. Enteric fever in Mediterranean north Africa. J. Infect. Dev. Ctries 2009, 3, 753–761. [Google Scholar]
- Srikantiah, P.; Girgis, F.Y.; Luby, S.P.; Jennings, G.; Wasfy, M.O.; Crump, J.A.; Hoekstra, R.M.; Anwer, M.; Mahoney, F.J. Population-based surveillance of typhoid fever in Egypt. Am. J. Trop. Med. Hyg 2006, 74, 114–119. [Google Scholar]
- Guard-Petter, J. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol 2001, 3, 421–430. [Google Scholar]
- Bautista, D.; Elankumaran, S.; Arking, J.; Heckert, R. Evaluation of an immunochromatography strip assay for the detection of Salmonella sp. from poultry. J. Vet. Diagn. Invest 2002, 14, 427–430. [Google Scholar]
- Gehring, A.G.; Gerald, C.; Mazenko, R.S.; Van Houten, L.J.; Brewster, J.D. Enzyme-linked immunomagnetic electrochemical detection of Salmonella typhimurium. J. Immunol. Methods 1996, 195, 15–25. [Google Scholar]
- Jain, S.; Chattopadhyay, S.; Jackeray, R.; Abid, C.K.V.Z.; Kohli, G.S.; Singh, H. Highly sensitive detection of Salmonella typhi using surface aminated polycarbonate membrane enhanced-ELISA. Biosens. Bioelectron 2012, 31, 37–43. [Google Scholar]
- Jaradat, Z.W.; Bzikot, J.H.; Zawistowski, J.; Bhunia, A.K. Optimization of a rapid dot-blot immunoassay for detection of Salmonella enterica serovar Enteritidis in poultry products and environmental samples. Food Microbiol 2004, 21, 761–769. [Google Scholar]
- Magliulo, M.; Simoni, P.; Guardigli, M.; Michelini, E.; Luciani, M.; Lelli, R.; Roda, A. A rapid multiplexed chemiluminescent immunoassay for the detection of Escherichia coli O157: H7, Yersinia enterocolitica, Salmonella typhimurium, and Listeria monocytogenes pathogen bacteria. J. Agric. Food Chem 2007, 55, 4933–4939. [Google Scholar]
- Nicholas, R.A.; Cullen, G.A. Development and application of an ELISA for detecting antibodies to Salmonella enteritidis in chicken flocks. Vet. Rec 1991, 128, 74–76. [Google Scholar]
- Preechakasedkit, P.; Pinwattana, K.; Dungchai, W.; Siangproh, W.; Chaicumpa, W.; Tongtawe, P.; Chailapakul, O. Development of a one-step immunochromatographic strip test using gold nanoparticles for the rapid detection of Salmonella typhi in human serum. Biosens. Bioelectron 2012, 31, 562–566. [Google Scholar]
- Proux, K.; Houdayer, C.; Humbert, F.; Cariolet, R.; Rose, V.; Eveno, E.; Madec, F. Development of a complete ELISA using Salmonella lipopolysaccharides of various serogroups allowing to detect all infected pigs. Vet. Res 2000, 31, 481–490. [Google Scholar]
- Malorny, B.; Bunge, C.; Helmuth, R. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 2007, 70, 245–251. [Google Scholar]
- Miller, N.D.; Davidson, P.M.; D’Souza, D.H. Real-time reverse-transcriptase PCR for Salmonella Typhimurium detection from lettuce and tomatoes. LWT-Food Sci. Technol 2011, 44, 1088–1097. [Google Scholar]
- Nam, H.M.; Srinivasan, V.; Gillespie, B.E.; Murinda, S.E.; Oliver, S.P. Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int. J. Food Microbiol 2005, 102, 161–171. [Google Scholar]
- Saroj, S.D.; Shashidhar, R.; Karani, M.; Bandekar, J.R. Rapid, sensitive, and validated method for detection of Salmonella in food by an enrichment broth culture-nested PCR combination assay. Mol. Cell. Probes 2008, 22, 201–206. [Google Scholar]
- Verdoy, D.; Barrenetxea, Z.; Berganzo, J.; Agirregabiria, M.; Ruano-Lopez, J.M.; Marimon, J.M.; Olabarria, C. A novel real time micro PCR based Point-of-Care device for Salmonella detection in human clinical samples. Biosens. Bioelectron 2012, 32, 259–265. [Google Scholar]
- Chen, W.; Xu, D.; Liu, L.; Peng, C.; Zhu, Y.; Ma, W.; Bian, A.; Li, Z.; Yuan, Y.; Jin, Z.; et al. Ultrasensitive detection of trace protein by western blot based on POLY-quantum dot probes. Anal. Chem 2009, 81, 9194–9198. [Google Scholar]
- Peng, C.; Li, Z.; Zhu, Y.; Chen, W.; Yuan, Y.; Liu, L.; Li, Q.; Xu, D.; Qiao, R.; Wang, L.; et al. Simultaneous and sensitive determination of multiplex chemical residues based on multicolor quantum dot probes. Biosens. Bioelectron 2009, 24, 3657–3662. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kuang, H.; Cui, G.; Chen, X.; Yin, H.; Yong, Q.; Xu, L.; Peng, C.; Wang, L.; Xu, C. A One-Step Homogeneous Sandwich Immunosensor for Salmonella Detection Based on Magnetic Nanoparticles (MNPs) and Quantum Dots (QDs). Int. J. Mol. Sci. 2013, 14, 8603-8610. https://doi.org/10.3390/ijms14048603
Kuang H, Cui G, Chen X, Yin H, Yong Q, Xu L, Peng C, Wang L, Xu C. A One-Step Homogeneous Sandwich Immunosensor for Salmonella Detection Based on Magnetic Nanoparticles (MNPs) and Quantum Dots (QDs). International Journal of Molecular Sciences. 2013; 14(4):8603-8610. https://doi.org/10.3390/ijms14048603
Chicago/Turabian StyleKuang, Hua, Gang Cui, Xiujin Chen, Honghong Yin, Qianqian Yong, Liguang Xu, Chifang Peng, Libing Wang, and Chuanlai Xu. 2013. "A One-Step Homogeneous Sandwich Immunosensor for Salmonella Detection Based on Magnetic Nanoparticles (MNPs) and Quantum Dots (QDs)" International Journal of Molecular Sciences 14, no. 4: 8603-8610. https://doi.org/10.3390/ijms14048603




