Genetic Analysis of HIV-1 Integrase Sequences from Treatment Naive Individuals in Northeastern South Africa
Abstract
:1. Introduction
2. Results and Discussion
2.1. Subjects’ Demographics and Resistance Mutations
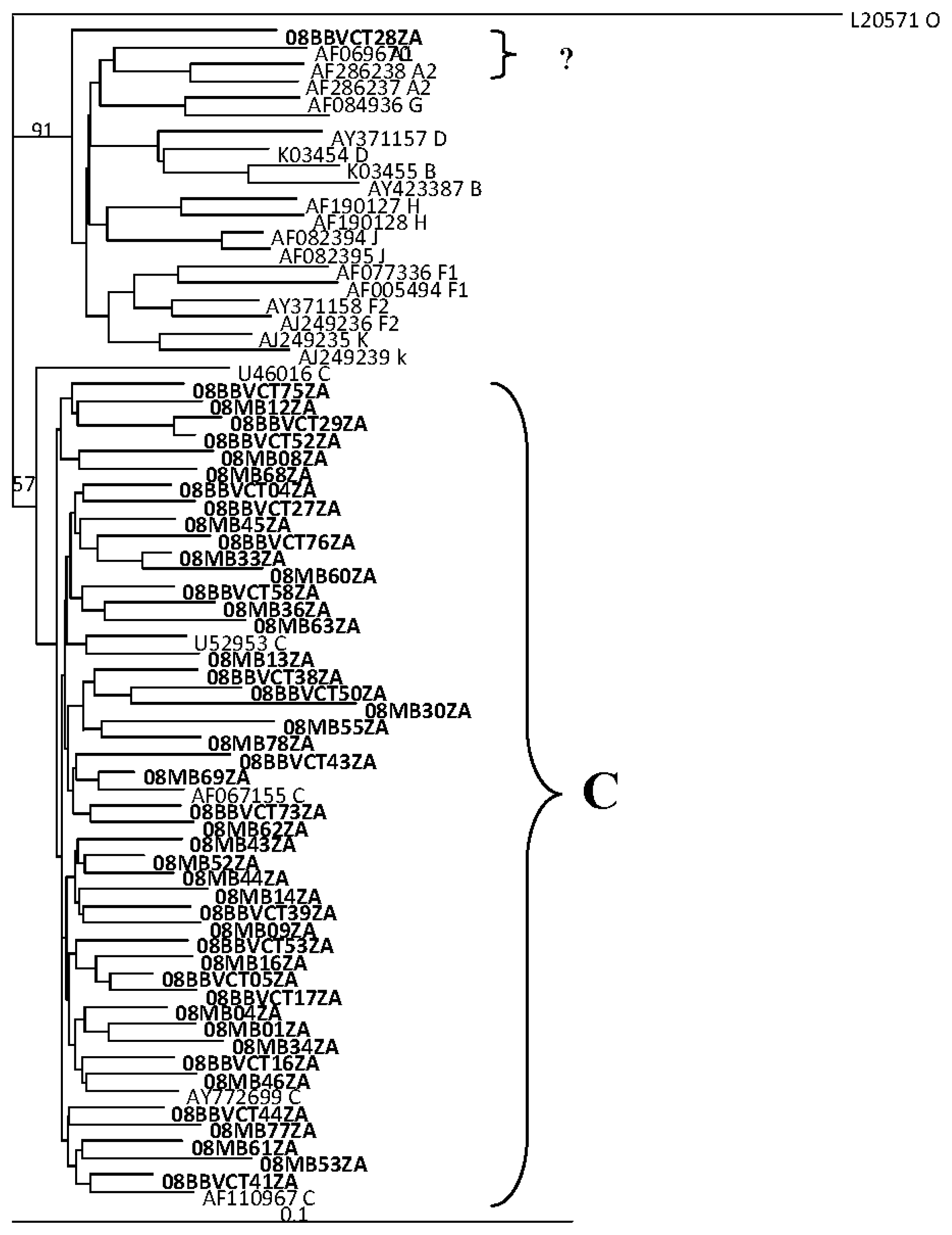
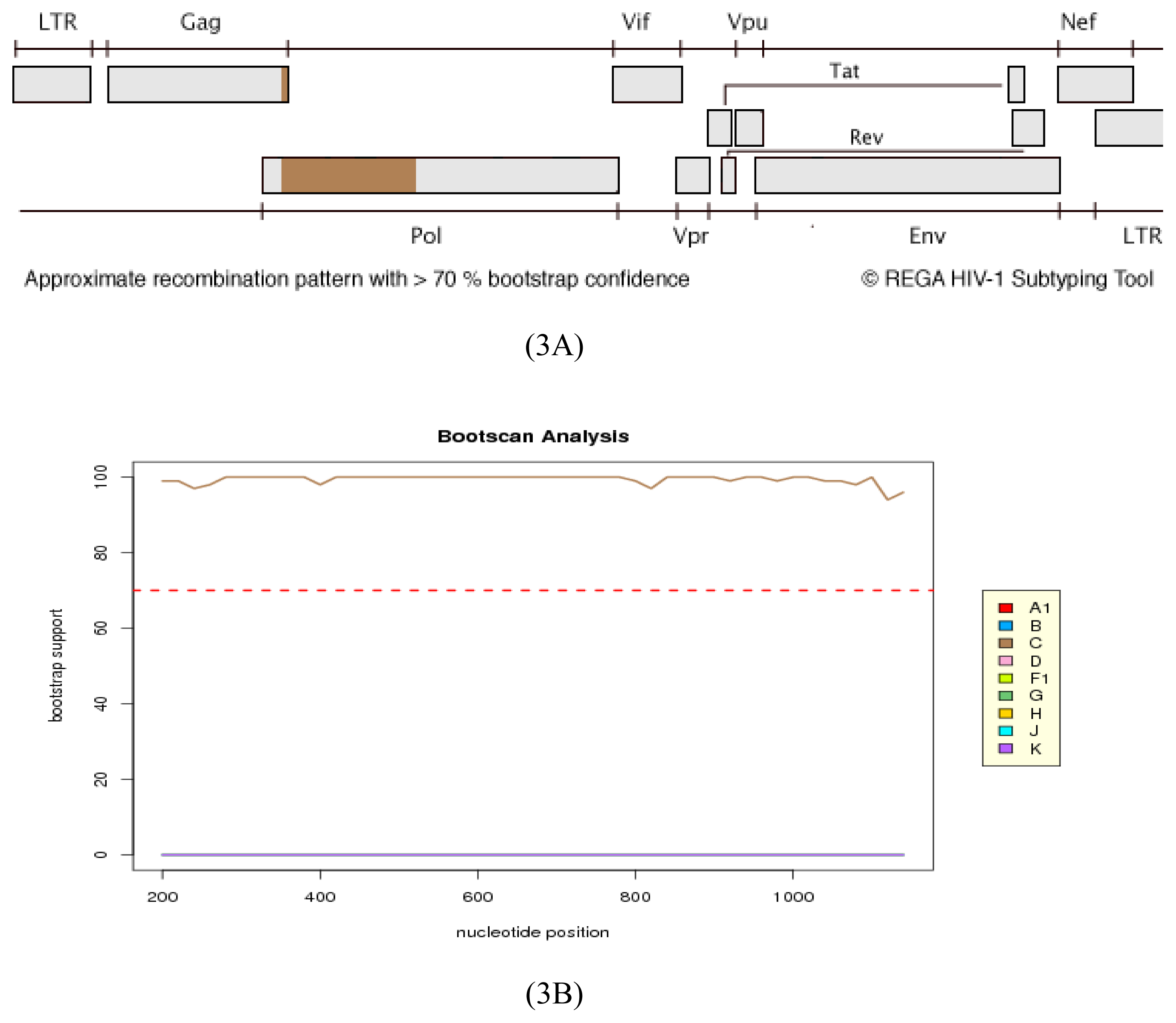
2.2. HIV Integrase Genotypes
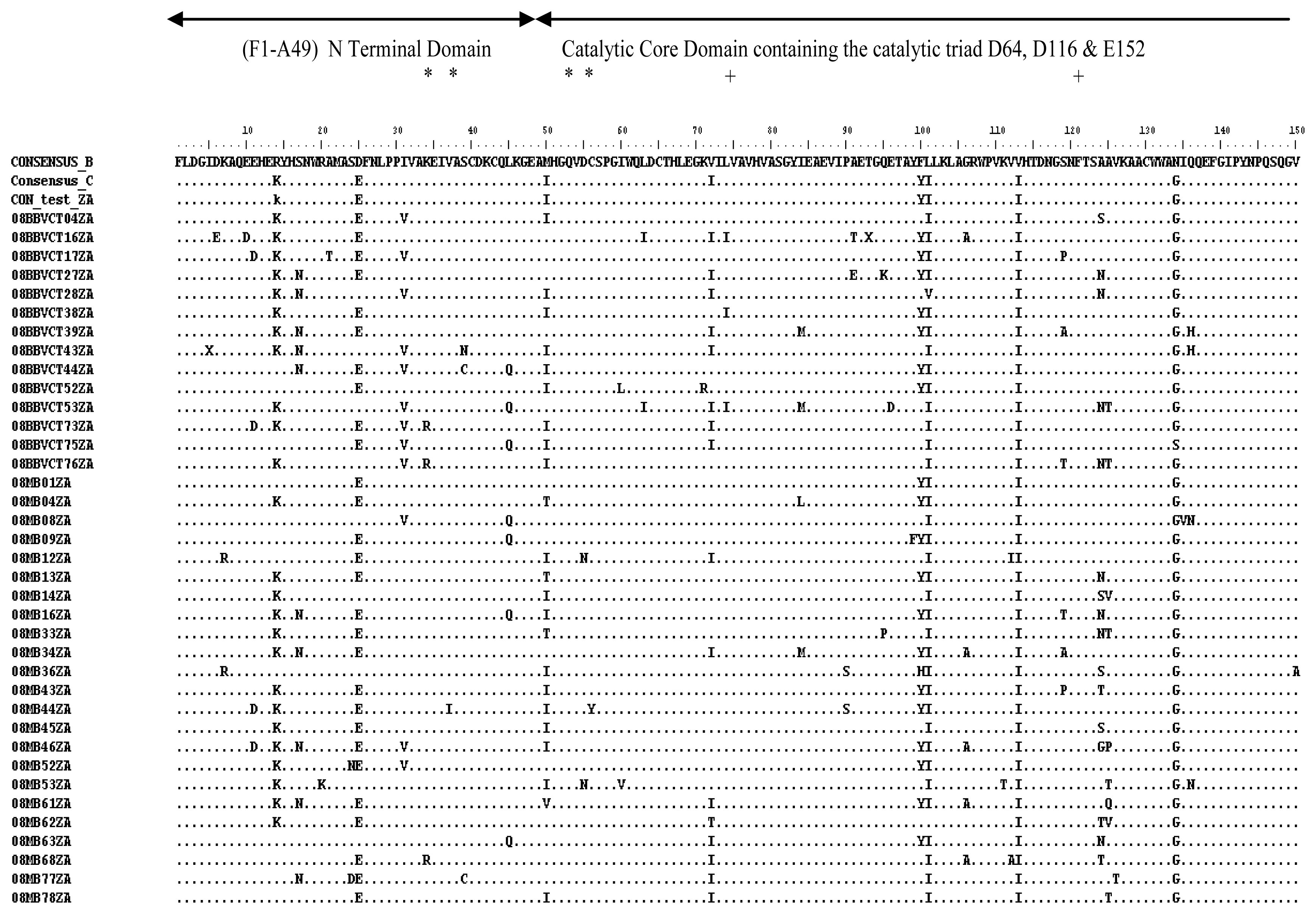
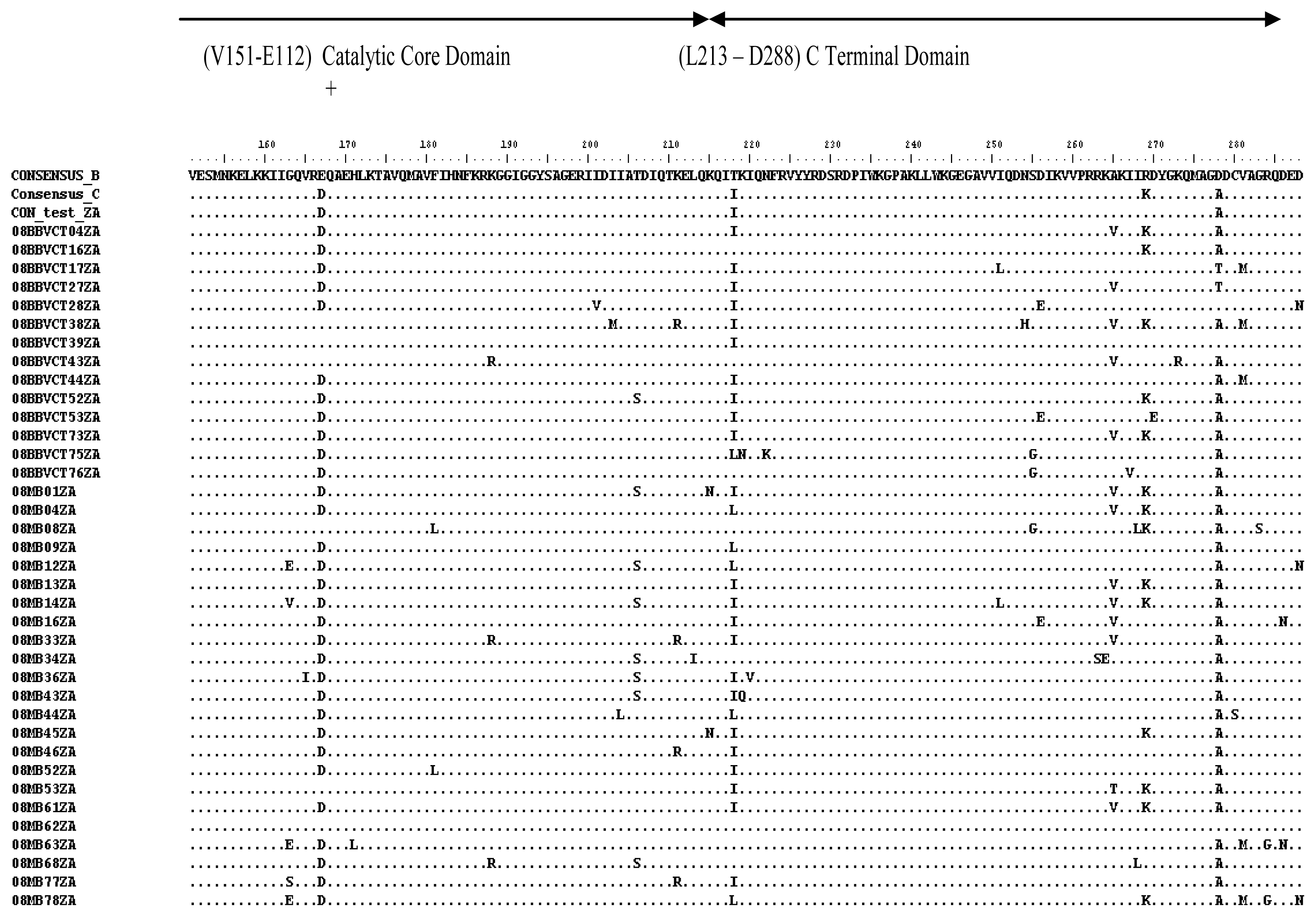
2.3. Analysis of the Functional Domains of HIV-1 IN Gene
3. Experimental Section
3.1. Ethical Considerations and Study Sites
3.2. Study Population, Sample Collection and Plasma Preparation
3.3. Viral RNA Isolation, RT-PCR and Nested PCR
3.4. Genetic Subtyping and Resistance Analyses
3.5. Analysis of the Functional Domains of HIV-1 Integrase Gene
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Goethals, O.; Clayton, R.; Ginderen, M.V.; Vereycken, I.; Wagemans, E.; Geluykens, P.; Dockx, K.; Strijbos, R.; Smits, V.; Vos, A.; et al. Resistance mutations in HIV-1 integrase selected with Elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J. Virol. 2008, 82, 10366–10374. [Google Scholar]
- Mckee, C.J.; Kessl, J.J.; Shkriabia, N.; Dar, M.J.; Engelman, A.; Kvaratskhelia, M. Dynamic modulation of HIV-1 integrase structure and function by cellular Lens Epithelium-derived Growth Factor (LEDGF) Protein. J. Biol. Chem 2008, 283, 31802–31812. [Google Scholar]
- Ceccherini-Silberstein, F.; Malet, I.; D’Arrigo, R.; Antinori, A.; Marcelin, A.G.; Perno, C.F. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev 2009, 11, 17–29. [Google Scholar]
- The National Antenatal Sentinel HIV and Syphilis Prevalence Survey in South Africa; National Department of Health: Pretoria, South Africa, 2010; pp. 1–72.
- Cooper, D.; Gatell, J.; Rocktroh, J.; Katlama, C.; Yeni, P.; Lazzarin, A.; Chen, J.; Isaaces, R.; Teppler, H.; Nguyen, B. For the BENHMRK-1 Study Group, 2007. Results of BENCHMRK-1, a phase III study evaluating the efficacy and safety of MK-0518, a novel HIV-1 integrase inhibitor, in patients with triple-class resistant virus. Proceedings of the 15th Conference on Retroviruses and Opportunistic Infections, Boston, MA, USA, 3–6 February 2008. Abstract 105aLB.
- Stellbrink, H.J. Raltegravir in the management of HIV-infected patients. Drug Des. Dev. Ther 2008, 2, 281–288. [Google Scholar]
- Marconi, V.C.; Sunpath, H.; Lu, Z.; Gordon, M.; Koranteng-Apeagyei, K.; Hampton, J.; Carpenter, S.; Giddy, J.; Ross, D.; Holst, H.; et al. Prevalence of HIV-1 Drug resistance after failure of a first highly active antiretroviral regimen in KwaZulu Natal, South Africa. Clin. Inf. Dis. 2008, 46, 1589–1597. [Google Scholar]
- Fox, M.P.; Cutsem, G.V.; Giddy, J.; Maskew, M.; Keiser, O.; Proszesky, H.; Wood, R.; Hernan, M.A.; Sterne, J.A.; Egger, M.; et al. IeDEA-SA Collaboration. Rates and predictors of failure of first-line antiretroviral therapy and switch to second-line ART in South Africa. J. Acquired Immune Defic. Syndr. 2012, 60, 428–437. [Google Scholar]
- Nglazi, M.D.; Kranzer, K.; Holele, P.; Kaplan, R.; Mark, D.; Jaspan, H.; Lawn, S.D.; Wood, R.; Bekker, L.G. Treatment outcomes in HIV-infected adolescents attending a community-based antiretroviral therapy clinic in South Africa. BMC Infect. Dis 2012, 12, 21. [Google Scholar] [Green Version]
- Fish, M.Q.; Hewer, R.; Wallis, C.L.; Venter, W.D.; Stevens, W.S.; Papathanasopoulos, M.A. Natural polymorphisms of integrase among HIV type 1-infected South African Patients. AIDS Res. Hum. Retroviruses 2010, 26, 489–493. [Google Scholar]
- Eshleman, S.H.; Hudelson, S.E.; Smith, E.; Hackett, J., Jr; Holzmayer, V.; Swanson, P.; Devare, S.G.; Marlowe, N. Analysis of pol integrase sequences in diverse HIV type 1 strains using a prototype genotyping assay. AIDS Res. Hum. Retroviruses 2009, 25, 343–345. [Google Scholar]
- Oliveira, M.F.; Ramalho, D.B.; Abreu, C.M.; Vubil, A.; Mabunda, N.; Ismael, N.; Francisco, C.; Jani, I.V.; Tanuri, A. Genetic diversity and naturally polymorphisms in HIV type 1 integrase isolates from Maputo, Mozambique: Implications for integrase inhibitors. AIDS Res. Hum. Retroviruses 2012, 12, 368. [Google Scholar]
- Arruda, L.B.; Fonseca, L.A.; Duarte, A.J.; Casseb, J. Genetic diversity on the integrase region of the pol gene among HIV type 1-infected patients naïve for integrase inhibitors in Sao Paulo city, Brazil. AIDS Res. Hum. Retroviruses 2010, 26, 105–107. [Google Scholar]
- Ayouba, A.; Aghokeng, A.; Bazin, B.; Brun-Vézinet, F.; Chaix, M.L.; Descamps, D.; Fleury, H.; Kane, C.T.; Lien, T.X.; Masquelier, B.; et al. Drug resistance mutations of HIV type 1 non-B viruses to integrase inhibitors in treatment-naive patients from sub-saharan countries and discordant interpretations. AIDS Res. Hum. Retroviruses 2012, 28, 1157–1160. [Google Scholar]
- Bessong, P.O.; Mphahlele, J.; Choge, I.A.; Obi, L.C.; Morris, L.; Hammarskjöld, M.; Rekosh, D.M. Resistance mutational analysis of HIV Type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res. Hum. Retroviruses 2006, 22, 1306–1312. [Google Scholar]
- Bessong, P.O.; Obi, C.L.; Cilliers, T.; Choge, I.; Phoswa, M.; Pillay, C. Characterization of human immunodeficiency virus type 1 from a previously unexplored region of South Africa with a high HIV prevalence. AIDS Res. Hums Retroviruses 2005, 21, 103–109. [Google Scholar]
- Papathanasopoulos, M.; Vardas, E.; Wallis, C.; Giashoff, R.; Butto, S.; Poli, G.; Malnati, M.; Clerici, M.; Ensoli, B. Characterization of HIV type 1 genetic diversity among South African participants enrolled in the AIDS Vaccine Integrated Project (AVIP) study. AIDS Res. Hum. Retroviruses 2010, 26, 705–709. [Google Scholar]
- Van den Ent, F.M.; Vos, A.; Plasterk, R.H. Mutational scan of the human immunodeficiency virus type 2 integrase protein. J. Virol 1998, 72, 3916–3924. [Google Scholar]
- Engelman, A.; Liu, Y.; Chen, H.; Farzan, M.; Dyda, F. Structure based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol 1997, 71, 3507–3514. [Google Scholar]
- Nwobegahay, J.; Selabe, G.; Ndjeka, N.O.; Manhaeve, C.; Bessong, P.O. Low prevalence of transmitted genetic drug resistance in a cohort of HIV infected naïve patients entering antiretroviral treatment programs at two sites in northern South Africa. J. Med. Virol 2012, 84, 1839–1843. [Google Scholar]
- Alcantara, L.C.; Cassol, S.; Libin, P.; Deforche, K.; Pybus, O.G.; van Ranst, M.; Galvão-Castro, B.; Vandamme, A.M.; de Oliveira, T. A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 2009, 10, 1–9. [Google Scholar]
- De Oliveira, T.; Deforche, K.; Cassol, S.; Salminen, M.; Paraskevis, D.; Seebregts, C.; Snoeck, J.; van Rensburg, E.J.; Wensing, A.M.J.; van de Vijver, D.A.; et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 2005, 21, 3797–3800. [Google Scholar]
- Stanford University HIV Drug Resistance Database. Available online: http://hivdb.stanford.edu accessed on 1 November 2012.
- Johnson, V.A.; Calvez, V.; Gunthard, H.F.; Paredes, R.; Pillay, D.; Shafer, R.; Wensing, A.M.; Richman, D.D. Update of the drug resistance mutations in HIV-1. Top. Antivir. Med 2011, 19, 156–164. [Google Scholar]
- Biological sequence alignment editor for windows 95/98/NT/XP/Vista/7. Available online: http://www.mbio.ncsu.edu/BioEdit/bioedit.html accessed on 1 November 2012.

| Sample code | Integrase mutation | Frequency | Coding nucleotides | Comment | |
|---|---|---|---|---|---|
| Wild type | mutant | ||||
| 08BBVCT73ZA | L74M | 1/89 (1.1%) | CTG | ATG | Polymorphism (Stanford); minor mutation involved in the Q148H/K/R pathway to Raltegravir resistance (IAS). |
| 08BBVCT27ZA | Q95K | 1/89 (1.1%) | CAA | AAA | Minor mutation to Raltegravir (Stanford); not documented in IAS algorithm |
| 08MB33ZA | Q95P | 1/89 (1.1%) | CAA | CCA | Unusual mutation (Stanford); not documented in IAS algorithm. |
| 08BBVCT49ZA | E157K | 1/89 (1.1%) | GAA | AAA | Unusual mutation (Stanford); mutation not documented in IAS algorithm. |
| 08BBVCT38ZA | I203M | 1/89 (1.1%) | ATA | ATG | Polymorphism; but not documented in IAS algorithm. |
| 08MB34Z | R263S | 1/89 (1.1%) | AGG | AGC | Unusual mutation by Stanford; but not documented in IAS algorithm. |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bessong, P.O.; Nwobegahay, J. Genetic Analysis of HIV-1 Integrase Sequences from Treatment Naive Individuals in Northeastern South Africa. Int. J. Mol. Sci. 2013, 14, 5013-5024. https://doi.org/10.3390/ijms14035013
Bessong PO, Nwobegahay J. Genetic Analysis of HIV-1 Integrase Sequences from Treatment Naive Individuals in Northeastern South Africa. International Journal of Molecular Sciences. 2013; 14(3):5013-5024. https://doi.org/10.3390/ijms14035013
Chicago/Turabian StyleBessong, Pascal Obong, and Julius Nwobegahay. 2013. "Genetic Analysis of HIV-1 Integrase Sequences from Treatment Naive Individuals in Northeastern South Africa" International Journal of Molecular Sciences 14, no. 3: 5013-5024. https://doi.org/10.3390/ijms14035013





