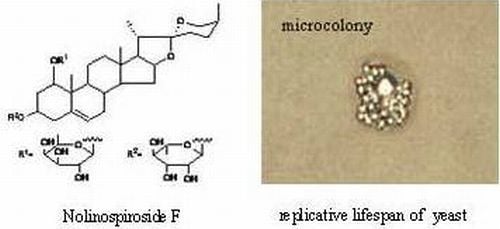
A Steroidal Saponin from Ophiopogon japonicus Extends the Lifespan of Yeast via the Pathway Involved in SOD and UTH1
Abstract
:Abbreviations
| AD | Alzheimer’s disease |
| MS | mass spectrometry |
| MDA | malondialdehyde |
| NMR | nuclear magnetic resonance |
| O. japonicus | Ophiopogon japonicus |
| RES | resveratrol |
| ROS | reactive oxygen species |
| RT-PCR | real-time polymerase chain reaction |
| SOD | superoxide dismutase |
| YPD | yeast peptone dextrose |
| YPGal | yeast peptone galactose |
1. Introduction
2. Results and Discussion
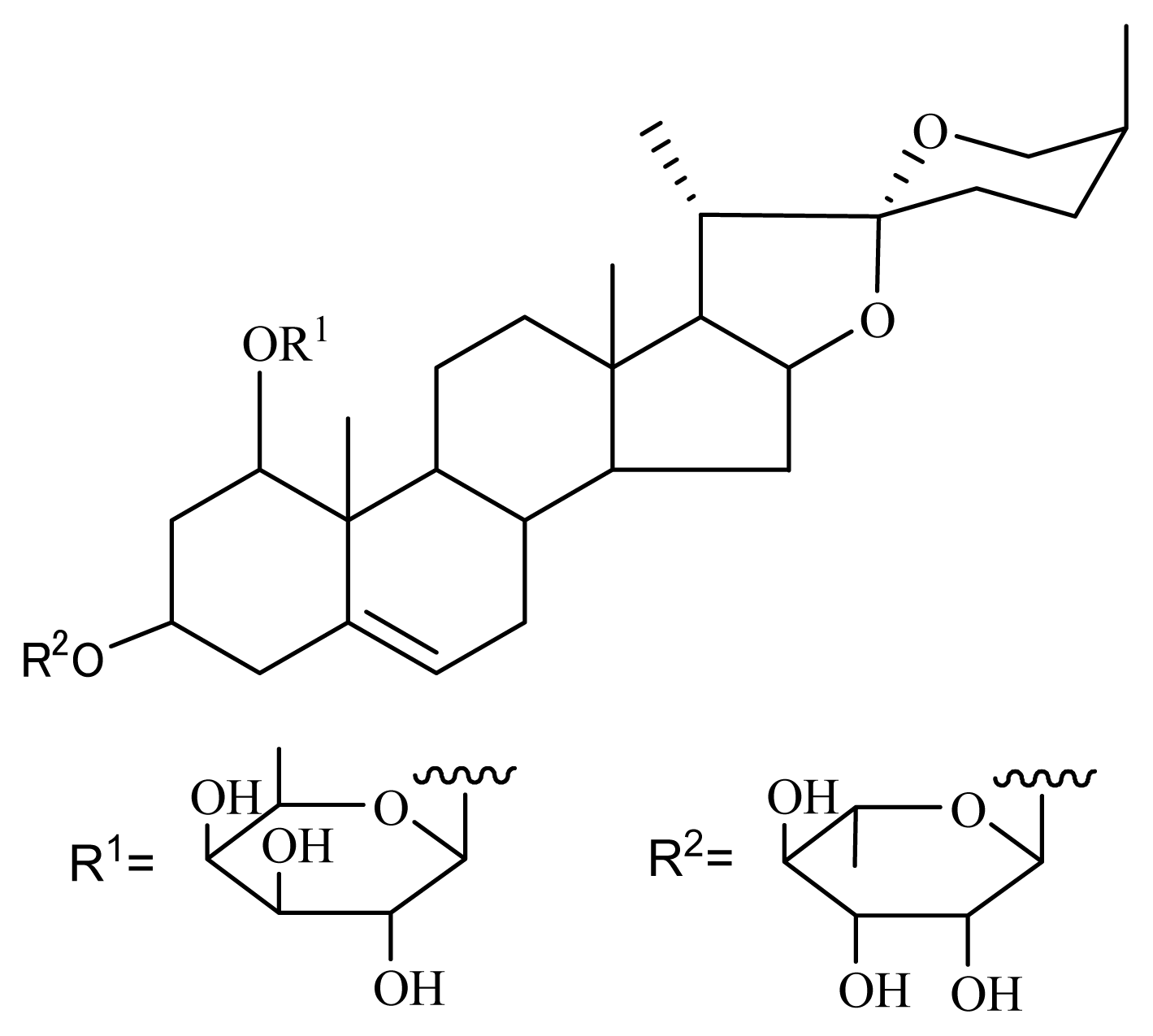
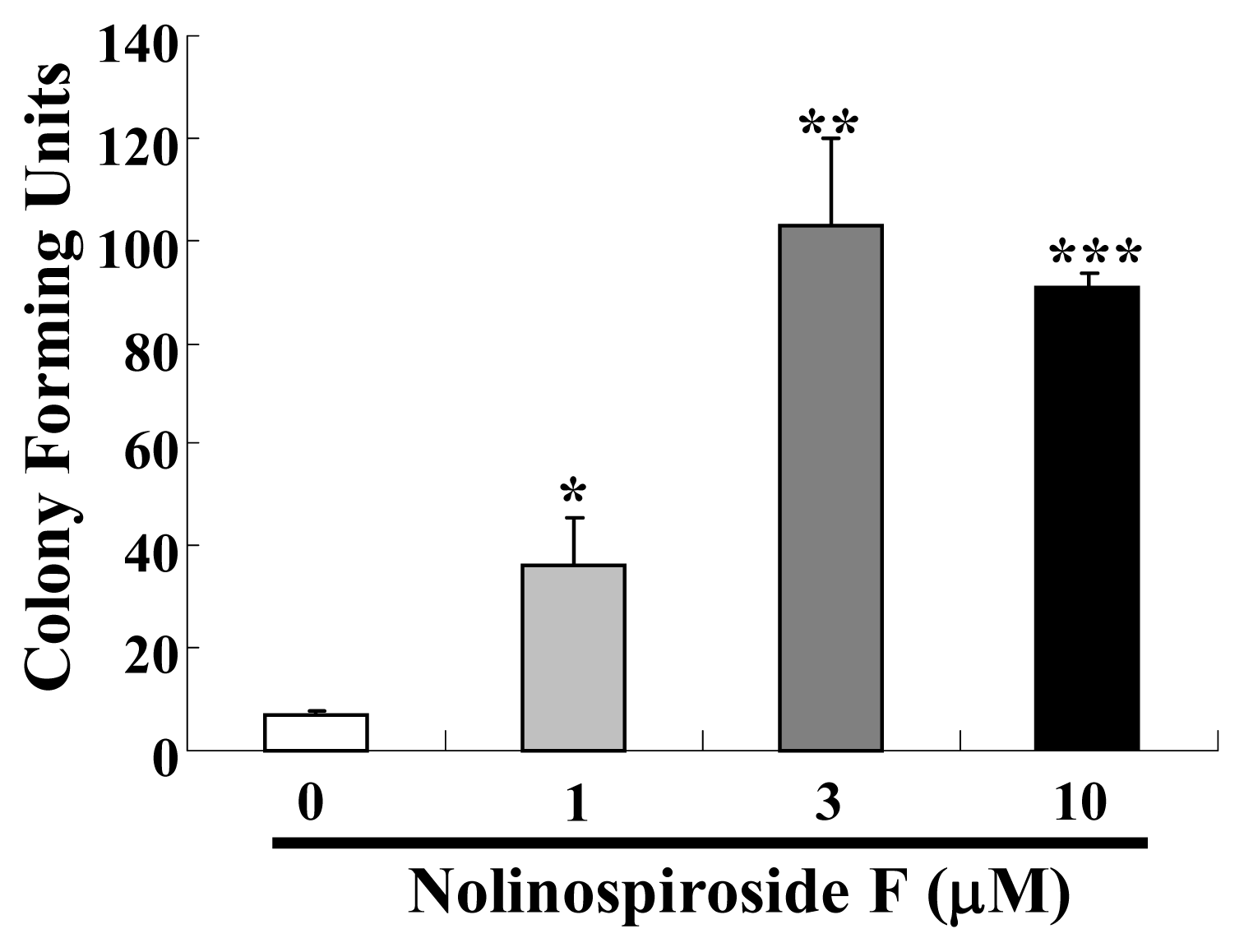
2.1. Nolinospiroside F Has Anti-Aging Effects in Yeast
2.2. Nolinospiroside F Affects the Growth of Yeast under Oxidative Stress Conditions
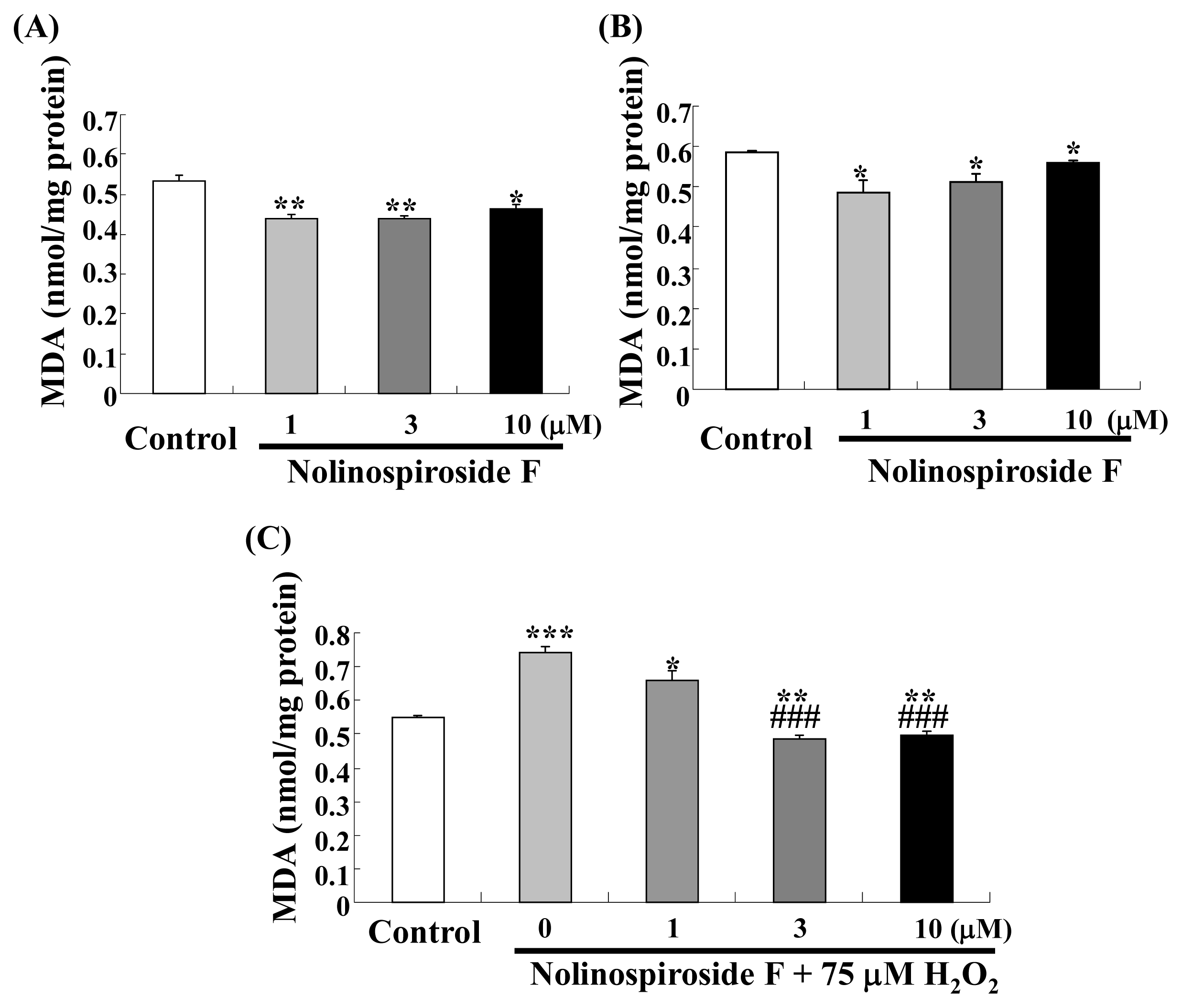
2.3. Nolinospiroside F Decreases MDA Levels
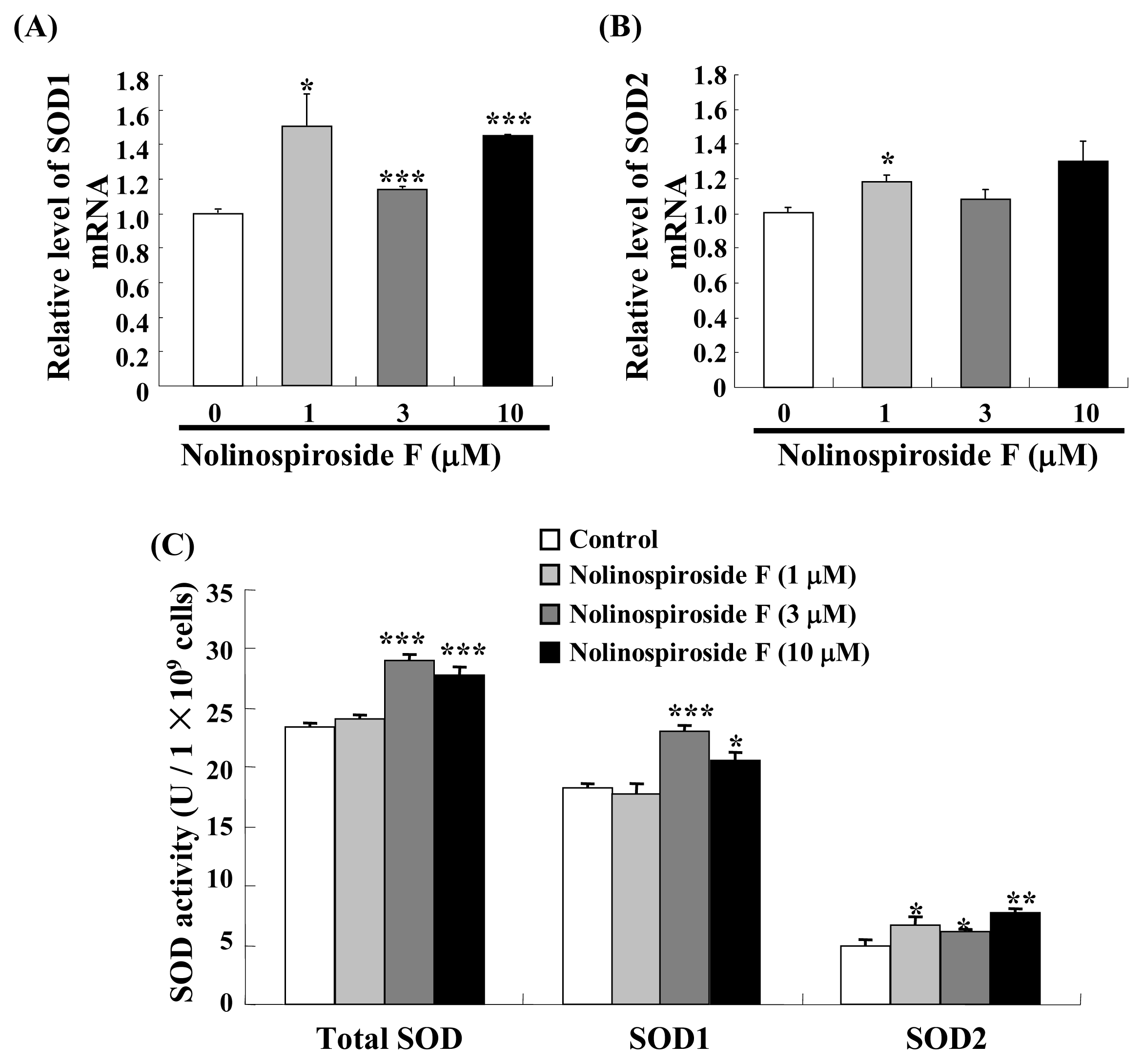
2.4. Treatment with Nolinospiroside F Increases SOD Gene Expression and SOD Enzyme Activity
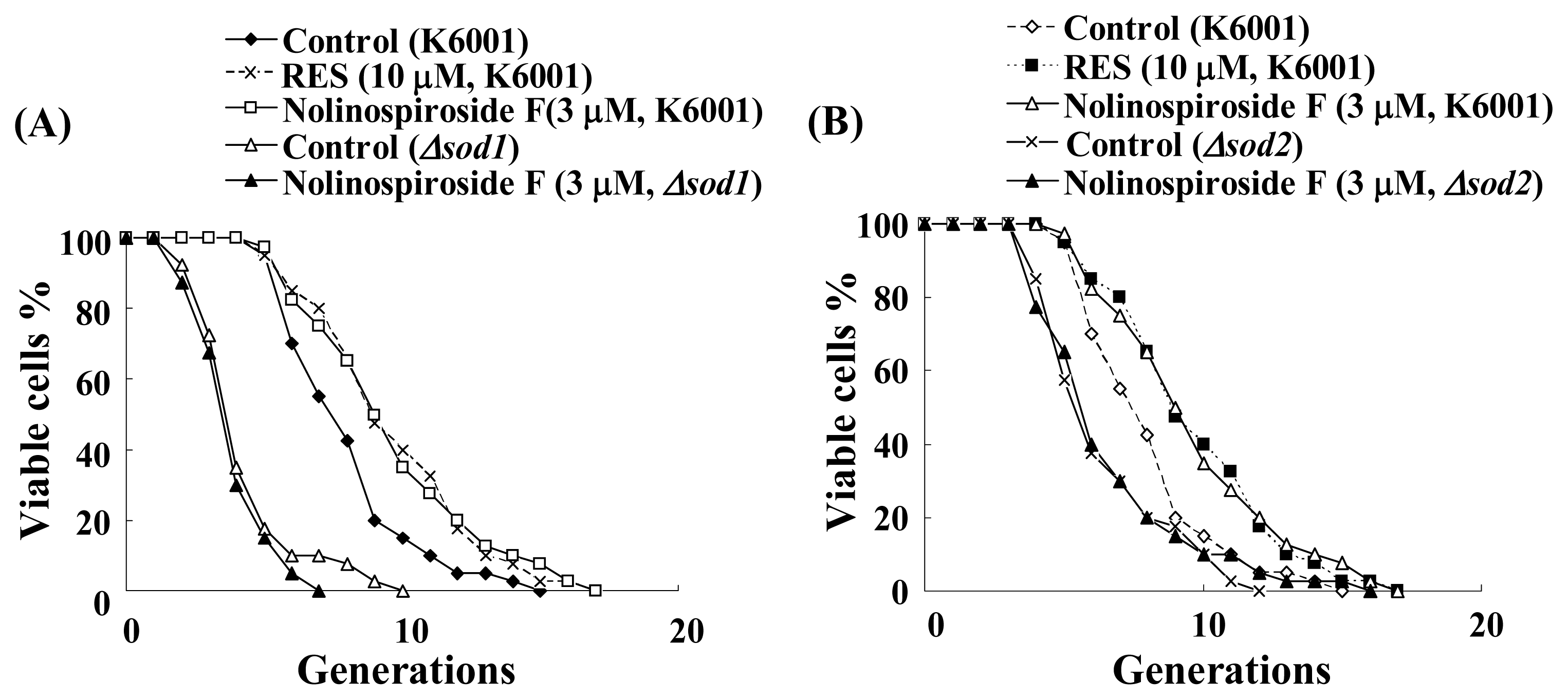
2.5. Nolinospiroside F Does Not Affect the Lifespans of sod1 and sod2 Yeast Mutant Strains
2.6. Nolinospiroside F Increased SIRT1 Activity
2.7. Nolinospiroside F Administration Inhibits UTH1 Gene Expression and UTH1, SKN7 Are Involved in Nolinospiroside F-Mediated Lifespan Extension
2.8. The Relationship between the SOD and UTH1 Gene
3. Materials and Methods
3.1. Isolation of Nolinospiroside F
3.2. Saccharomyces cerevisiae Strains, Media and Lifespan Assay
3.3. Anti-Oxidative Stress Assay
3.4. MDA Assay
3.5. RT-PCR Analysis
3.6. SOD Activity Assay
3.7. SIRT1 Assay
3.8. Statistical Analysis
4. Conclusion
Acknowledgments
Conflict of Interest
References
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006, 444, 337–342. [Google Scholar]
- Parker, J.A.; Arango, M.; Abderrahmane, S.; Lambert, E.; Tourette, C.; Catoire, H.; Néri, C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet 2005, 37, 349–350. [Google Scholar]
- Araki, T.; Sasaki, Y.; Milbrandt, J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 2004, 305, 1010–1013. [Google Scholar]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar]
- Pasinetti, G.M.; Wang, J.; Marambaud, P.; Ferruzzi, M.; Gregor, P.; Knable, L.A.; Ho, L. Neuroprotective and metabolic effects of resveratrol: Therapeutic implications for Huntington’s disease and other neurodegenerative disorders. Exp. Neurol 2011, 232, 1–6. [Google Scholar]
- Shiozaki, M.; Hayakawa, N.; Shibata, M.; Koike, M.; Uchiyama, Y.; Gotow, T. Closer association of mitochondria with lipid droplets in hepatocytes and activation of KupVer cells in resveratrol-treated senescence-accelerated mice. Histochem. Cell Biol 2011, 136, 475–489. [Google Scholar]
- Valenzano, D.R.; Terzibasi, E.; Genade, T.; Cattaneo, A.; Domenici, L.; Cellerino, A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol 2006, 16, 296–300. [Google Scholar]
- Jarolim, S.; Millen, J.; Heeren, G.; Laun, P.; Goldfarb, D.S.; Breitenbach, M. A novel assay for replicative lifespan in Saccharomyces cerevisiae. FEMS Yeast Res 2004, 5, 169–177. [Google Scholar]
- Weng, Y.; Xiang, L.; Matsuura, A.; Zhang, Y.; Huang, Q.; Qi, J. Ganodermasides A and B, two novel anti-aging ergosterols from spores of a medicinal mushroom Ganoderma lucidum on yeast via UTH1 gene. Bioorg. Med. Chem 2010, 18, 999–1002. [Google Scholar]
- Weng, Y.; Lu, J.; Xiang, L.; Matsuura, A.; Zhang, Y.; Huang, Q.; Qi, J. Ganodermasides C and D, two new anti-aging ergosterols from the spores of a medicinal mushroom Ganoderma lucidum. Biosci. Biotechnol. Biochem 2011, 75, 800–803. [Google Scholar]
- Xiang, L.; Sun, K.; Lu, J.; Weng, Y.; Taoka, A.; Sakagami, Y.; Qi, J. Anti-aging effects of phloridzin, an apple polyphenol, on yeast via SOD and Sir2 genes. Biosci. Biotechnol. Biochem 2011, 75, 854–858. [Google Scholar]
- Sun, K.; Xiang, L.; Ishihara, S.; Matsuura, A.; Sakagami, Y.; Qi, J. Anti-aging effects of hesperidin on Saccharomyces cerevisiae via inhibition of reactive oxygen species and UTH1 gene expression. Biosci. Biotechnol. Biochem 2012, 76, 640–645. [Google Scholar]
- Shevchuk, G.V.; Vollerner, Y.S.; Shashkov, A.S.; Chirva, V.Y. Steroids of the furostan and spirostan series from nolina microcarpa II. Structures of nolinospiroside D and nolinofurosides D, E, and F. Chem. Nat. Compd 1991, 5, 678–686. [Google Scholar]
- Qi, J.; Ojika, M.; Sakagami, Y. Linckosides A and B, two new neuritogenic steroid glycosides from the Okinawan starfish Linckia laevigata. Bioorg. Med. Chem 2002, 10, 1961–1966. [Google Scholar]
- Qi, J.; Ojika, M.; Sakagami, Y. Linckosides C–E, three new neuritogenic steroid glycosides from the Okinawan starfish Linckia laevigata. Bioorg. Med. Chem 2004, 12, 4259–4265. [Google Scholar]
- Han, C.; Qi, J.; Ojika, M. Structure-activity relationships of novel neuritogenic steroid glycosides from the Okinawan starfish Linckia laevigata. Bioorg. Med. Chem 2006, 14, 4458–4465. [Google Scholar]
- Hua, Z.; Carcache, D.A.; Tian, Y.; Li, Y.M.; Danishefsky, S.J. The synthesis and preliminary biological evaluation of a novel steroid with neurotrophic activity: NGA0187. J. Org. Chem 2005, 70, 9849–9856. [Google Scholar]
- Furuya, S.; Takayama, F.; Mimaki, Y.; Sashida, Y.; Satoh, K.; Sakagami, H. Cytotoxic activity of saponins from Camassia leichtlinii against human oral tumor cell lines. Anticancer Res 2001, 21, 959–964. [Google Scholar]
- Shao, B.; Guo, H.Z.; Cui, Y.J.; Ye, M.; Han, J.; Guo, D. Steroidal saponins from Smilax china and their anti-inflammatory activities. Phytochemistry 2007, 68, 623–630. [Google Scholar]
- Li, H.; Huang, W.; Wen, Y.Q.; Gong, G.H.; Zhao, Q.B.; Yu, G. Anti-thrombotic activity and chemical characterization of steroidal saponins from Dioscorea zingiberensis C.H. Wright. Fitoterapia 2010, 81, 1147–1156. [Google Scholar]
- Zhang, Y.; Zhang, Y.J.; Jacob, M.R.; Li, X.C.; Yang, C.R. Steroidal saponins from the stem of Yucca elephantipes. Phytochemistry 2008, 69, 264–270. [Google Scholar]
- Watanabe, Y.; Sanada, S.; Ida, Y.; Shoji, J. Comparative studies on the constituents of ophiopogonis tuber and its congeners. I. Studies of the constituents of the subterranean part of Liriope platyphylla Wang et Tang. (1). Chem. Pharm. Bull 1983, 31, 1980–1990. [Google Scholar]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol 1956, 11, 298–300. [Google Scholar]
- Harman, D. Origin and evolution of the free radical theory of aging: A brief personal history, 1954–2009. Biogerontology 2009, 10, 773–781. [Google Scholar]
- Pryor, W.A.; Stanley, J.P. Letter: A suggested mechanism for the production of malondialdehyde during the autoxidation of polyunsaturated fatty acids. Nonenzymatic production of prostaglandin endoperoxides during autoxidation. J. Org. Chem 1975, 40, 3615–3617. [Google Scholar]
- Debouzy, J.C.; Fauvelle, F.; Vezin, H.; Brasme, B.; Chancerelle, Y. Interaction of the malondialdehyde molecule with membranes. A differential scanning calorimetry, 1H-, 31P-NMR and ESR study. Biochem. Pharmacol 1992, 44, 1787–1793. [Google Scholar]
- Ohyashiki, T.; Sakata, N.; Matsui, K. A decrease of lipid fluidity of the porcine intestinal brush-border membranes by treatment with malondialdehyde. J. Biochem 1992, 111, 419–423. [Google Scholar]
- Yang, M.; Schaich, K.M. Factors affecting DNA damage caused by lipid hydroperoxides and aldehydes. Free Radic. Biol. Med 1996, 20, 225–236. [Google Scholar]
- Kim, Y.H.; Park, K.H.; Rho, H.M. Transcriptional activation of the Cu, Zn-superoxide dismutase gene through the AP2 site by ginsenoside Rb2 extracted from a medicinal plant, Panax ginseng. J. Biol. Chem 1996, 271, 24539–24543. [Google Scholar]
- St.-Pierre, J.; Drori, S.; Uldry, M.; Silvaggi, J.M.; Rhee, J.; Jager, S.; Handschin, C.; Zheng, K.; Lin, J.; Yang, W.; et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1α transcriptional coactivators. Cell 2006, 127, 397–408. [Google Scholar]
- Daitoku, H.; Hatta, M.; Matsuzaki, H.; Aratani, S.; Ohshima, T.; Miyagishi, M.; Nakajima, T.; Fukamizu, A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10042–10047. [Google Scholar]
- Tanno, M.; Kuno, A.; Yano, T.; Miura, T.; Hisahara, S.; Ishikawa, S.; Shimamoto, K.; Horio, Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J. Biol. Chem 2010, 285, 8375–8382. [Google Scholar]
- Camougrand, N.; Kissová, I.; Velours, G.; Manon, S. Uth1p: A yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res 2004, 5, 133–140. [Google Scholar]
- Austriaco, N.R., Jr. Review: To bud until death: The genetics of ageing in the yeast Saccharomyces. Yeast 1996, 12, 623–630. [Google Scholar]
- Kennedy, B.K.; Austriaco, N.R., Jr; Zhang, J.; Guarente, L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 1995, 80, 485–496. [Google Scholar]
- Bandara, P.D.; Flattery-O’Brien, J.A.; Grant, C.M.; Dawes, I.W. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr. Genet 1998, 34, 259–268. [Google Scholar]




© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sun, K.; Cao, S.; Pei, L.; Matsuura, A.; Xiang, L.; Qi, J. A Steroidal Saponin from Ophiopogon japonicus Extends the Lifespan of Yeast via the Pathway Involved in SOD and UTH1. Int. J. Mol. Sci. 2013, 14, 4461-4475. https://doi.org/10.3390/ijms14034461
Sun K, Cao S, Pei L, Matsuura A, Xiang L, Qi J. A Steroidal Saponin from Ophiopogon japonicus Extends the Lifespan of Yeast via the Pathway Involved in SOD and UTH1. International Journal of Molecular Sciences. 2013; 14(3):4461-4475. https://doi.org/10.3390/ijms14034461
Chicago/Turabian StyleSun, Kaiyue, Shining Cao, Liang Pei, Akira Matsuura, Lan Xiang, and Jianhua Qi. 2013. "A Steroidal Saponin from Ophiopogon japonicus Extends the Lifespan of Yeast via the Pathway Involved in SOD and UTH1" International Journal of Molecular Sciences 14, no. 3: 4461-4475. https://doi.org/10.3390/ijms14034461