Expression Pattern of Class B Gene PAP3 in Flower Development of Pepper
Abstract
:1. Introduction
2. Results
2.1. Comparison with Anther Transcriptome
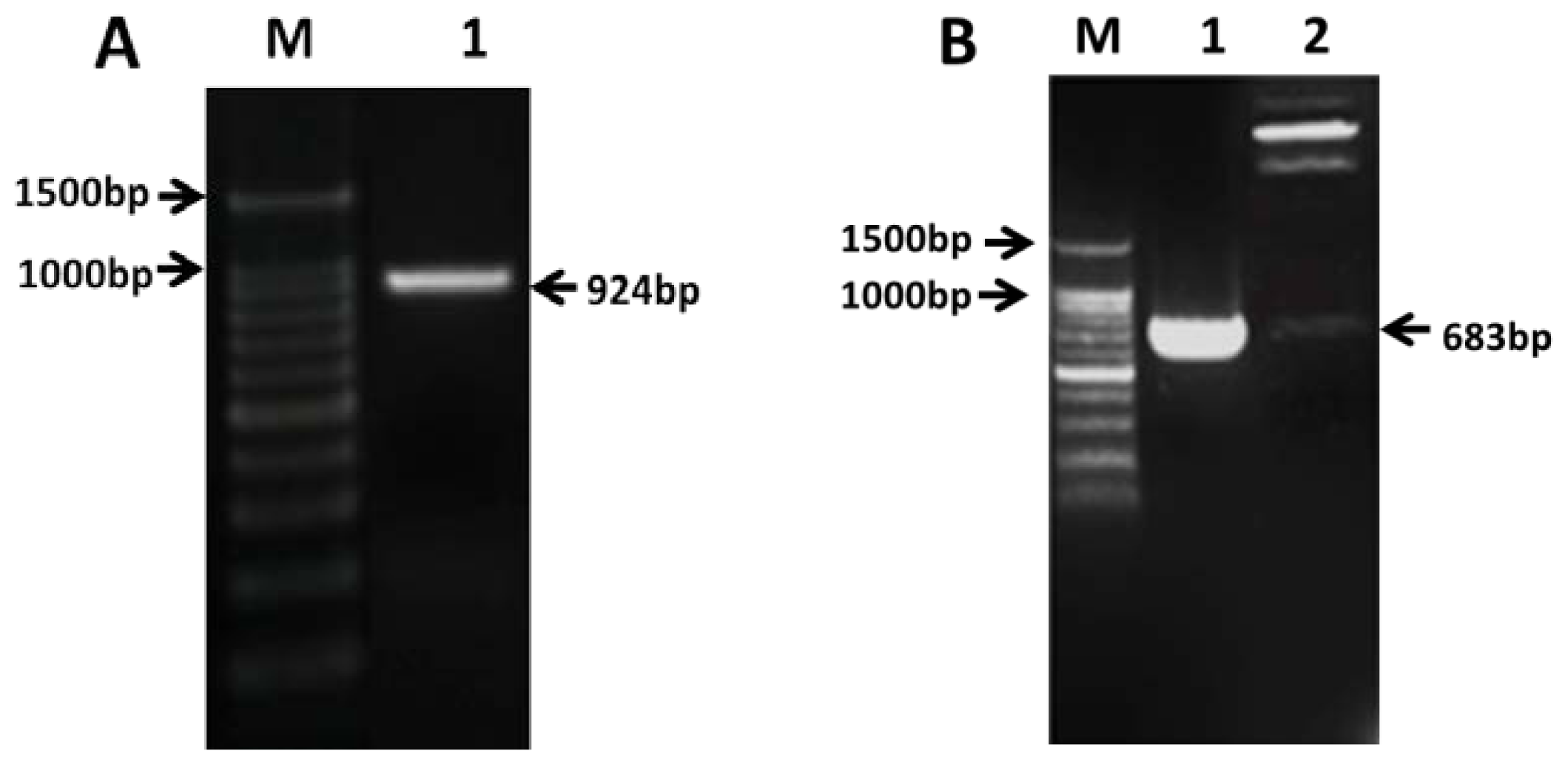
2.2. Cloning of PAP3 in CMS Line 121A
2.3. Construction of Transient Expressing Vector
2.4. Subcellular Localization of Gene Expression
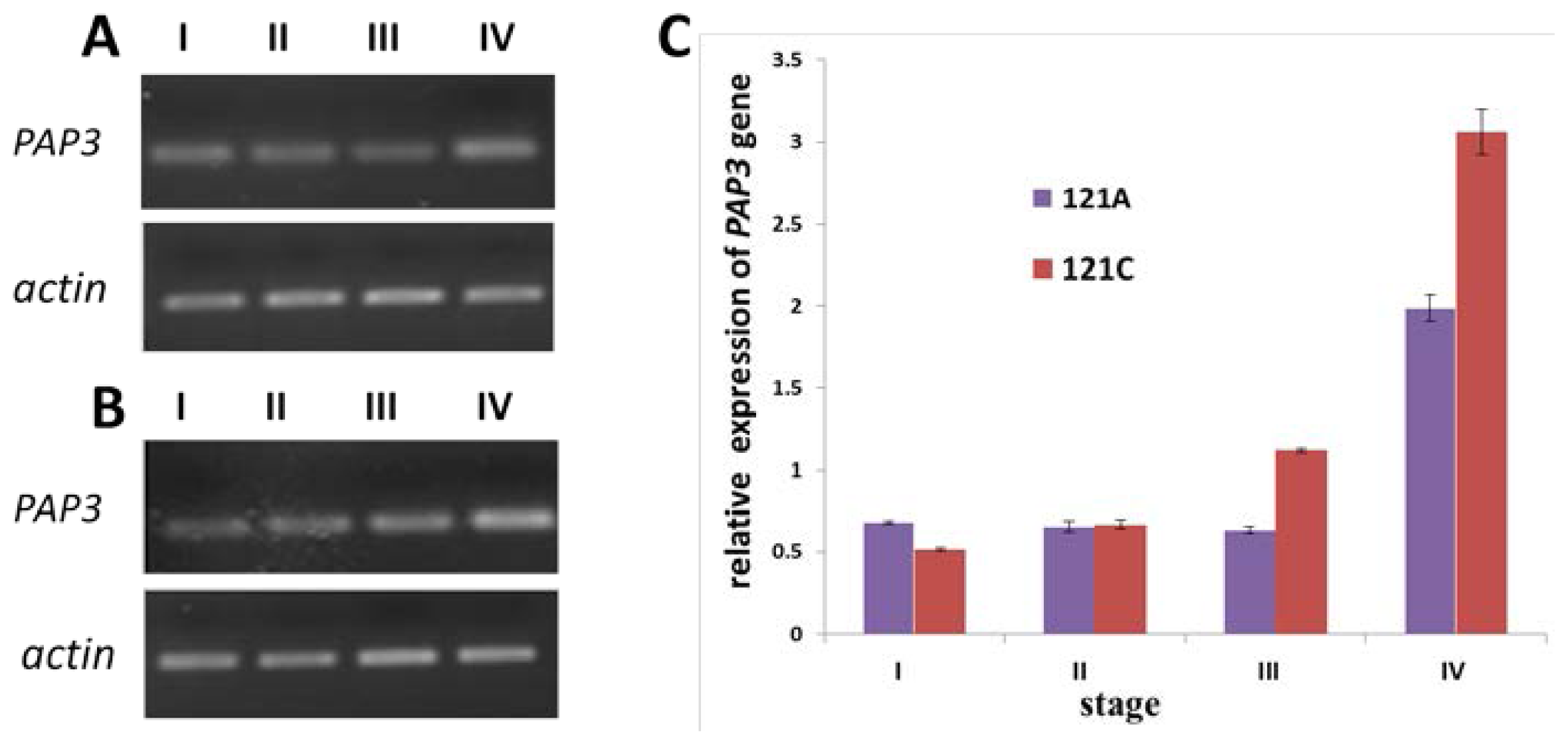
2.5. Expression of PAP3 Measured by Semi-Quantitative RT-PCR and qRT-PCR
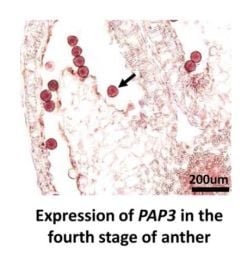
2.6. RNA in Situ Hybridization of PAP3
3. Discussion
4. Experimental Section
4.1. Materials of Plants
4.2. Gene Cloning and Blast
4.3. RNA Extraction and cDNA Synthesis
4.4. Cloning of PAP3 in CMS Line
4.5. Transient Expression of PAP3 in Onion Epidermal Cells
4.6. Semi Quantitative RT-PCR and qRT-PCR
4.7. In Situ Hybrization
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mackenzie, S.A. The influence of mitochondrial genetics on crop breeding strategies. Plant Breed. Rev 2005, 25, 115–138. [Google Scholar]
- Schnable, P.S.; Wise, R.P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 1998, 3, 175–180. [Google Scholar]
- Liu, W.Y.; Gniffke, P.A. Stability of AVRDC’s cytoplasmic male sterile (CMS) pepper lines grown under low temperature. Capsicum Eggplant Newsl 2004, 23, 85–88. [Google Scholar]
- Zhang, B.X.; Huang, S.W.; Yang, G.; Guo, J. Two RAPD markers linked to a major fertility restorer gene in pepper. Euphytica 2000, 113, 155–161. [Google Scholar]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet 2007, 23, 81–90. [Google Scholar]
- Linke, B.; Borner, T. Mitochondrial effects on flower and pollen development. Mitochondrion 2005, 5, 389–402. [Google Scholar]
- Carlsson, J.; Leino, M.; Sohlberg, J.; Sundstroem, J.F.; Glimelius, K. Mitochondrial regulation of flower development. Mitochondrion 2008, 8, 74–86. [Google Scholar]
- Gulyas, G.; Shin, Y.S.; Kim, H.T.; Lee, J.S.; Hirata, Y. Altered transcript reveals an Orf507 sterility-related gene in chili pepper (Capsicum annuum L.). Plant Mol. Biol. Rep 2010, 28, 605–612. [Google Scholar]
- Liu, C.; Ma, N.; Wang, P.Y.; Nan, F.; Shen, H.L. Transcriptome sequencing and de novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.). PLoS One 2013, 8, e65209. [Google Scholar]
- Guo, S.; Shen, H.L.; Yang, W.C. Isolation of fertility restoration-related ESTs in pepper cytoplasmic male sterility lines using SSH. Acta Hortic. Sin 2009, 36, 1443–1449. [Google Scholar]
- Guo, S.; Ma, N.; Yang, W.C. Expression analysis of restorer alleles-induced genes in pepper. Agric. Sci. China 2011, 10, 1010–1015. [Google Scholar]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar]
- Purugganan, M.D.; Rounsley, S.D.; Schmidt, R.J.; Yanofsky, M.F. Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics 1995, 140, 345–356. [Google Scholar]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS-box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol 2001, 4, 75–85. [Google Scholar]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar]
- Litt, A.; Kramer, E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol 2010, 21, 129–137. [Google Scholar]
- Theissen, G.; Becker, A.; di Rosa, A.; Kanno, A.; Kim, J.T.; Munster, T.; Winter, K.U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol 2000, 42, 115–149. [Google Scholar]
- Jack, T.; Fox, G.L.; Meyerowitz, E.M. Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 1994, 76, 703–716. [Google Scholar]
- Lohmann, J.U.; Weigel, D. Building beauty: The genetic control of floral patterning. Dev. Cell 2002, 2, 135–142. [Google Scholar]
- Lamb, R.S.; Hill, T.A.; Tan, Q.K.; Irish, V.F. Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 2002, 129, 2079–2086. [Google Scholar]
- Laufs, P.; Coen, E.; Kronenberger, J.; Traas, J.; Doonan, J. Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 2003, 130, 785–796. [Google Scholar]
- Zhou, L.; Zhou, Y.T.; Wang, M.L.; Wang, H.Y.; Zhao, Y. Expressions and dimerization affinities of three highly identical APETALA3 genes in Brassica napus. Biol. Plant 2010, 54, 33–40. [Google Scholar]
- Roque, E.; Serwatowska, J.; Cruz Rochina, M.; Wen, J.Q.; Mysore, K.S.; Yenush, L.; Beltran, J.P.; Canas, L.A. Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J 2013, 73, 663–675. [Google Scholar]
- Zhang, Y.F.; Wang, X.F.; Zhang, W.X.; Yu, F.; Tian, J.H.; Li, D.R.; Guo, A.G. Functional analysis of the two Brassica AP3 genes involved in apetalous and stamen carpelloid phenotypes. PLoS One 2011, 6, e20930. [Google Scholar]
- Xiao, H.; Wang, Y.; Liu, D.F.; Wang, W.M.; Li, X.B.; Zhao, X.F.; Xu, J.C.; Zhai, W.X.; Zhu, L.H. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol 2003, 52, 957–966. [Google Scholar]
- Kyozuka, J.; Kobayashi, T.; Morita, M.; Shimamoto, K. Spatially and temporally regulated expression of rice MADS-box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 2000, 41, 710–718. [Google Scholar]
- Kang, H.G.; Jeon, J.S.; Lee, S.; An, G. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol 1998, 38, 1021–1029. [Google Scholar]
- Sharma, B.; Kramer, E. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae). New Phytol 2013, 197, 949–957. [Google Scholar]
- Sato, H.; Yoshida, K.; Mitsuda, N.; Ohme-Takagi, M.; Takamizo, T. Male-sterile and cleistogamous phenotypes in tall fescue induced by chimeric repressors of SUPERWOMAN1 and OsMADS58. Plant Sci 2012, 183, 183–189. [Google Scholar]
- Su, K.M.; Zhao, S.H.; Shan, H.Y.; Kong, H.Z.; Lu, W.L.; Theissen, G.; Chen, Z.D.; Meng, Z. The MIK region rather than the C-terminal domain of AP3-like class B floral homeotic proteins determines functional specificity in the development and evolution of petals. New Phytol 2008, 178, 544–558. [Google Scholar]
- Whipple, C.J.; Ciceri, P.; Padilla, C.M.; Ambrose, B.A.; Bandong, S.L.; Schmidt, R.J. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 2004, 131, 6083–6091. [Google Scholar]
- Pylatuik, J.D.; Lindsay, D.L.; Davis, A.R.; Bonham-Smith, P.C. Isolation and characterization of a Brassica napus cDNA corresponding to a B-class floral development gene. J. Exp. Bot 2003, 54, 2385–2387. [Google Scholar]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar]
- Teixeira, R.T.; Farbos, I.; Glimelius, K. Expression levels of meristem identity and homeotic genes are modified by nuclear-mitochondrial interactions in alloplasmic male-sterile lines of Brassica napus. Plant J 2005, 42, 731–742. [Google Scholar]
- Farbos, I.; Mouras, A.; Bereterbide, A.; Glimelius, K. Defective cell proliferation in the floral meristem of alloplasmic plants of Nicotiana tabacum leads to abnormal floral organ development and male sterility. Plant J 2001, 26, 131–142. [Google Scholar]
- Kofer, W.; Glimelius, K.; Bonnett, H.T. Modifications of mitochondrial DNA cause changes in floral development in homeotic-like mutants of tobacco. Plant Cell 1991, 3, 759–769. [Google Scholar]
- Linke, B.; Nothnagel, T.; Borner, T. Flower development in carrot CMS plants: Mitochondria affect the expression of MADS-box genes homologous to GLOBOSA and DEFICIENS. Plant J 2003, 34, 27–37. [Google Scholar]
- Leino, M.; Teixeira, R.; Landgren, M.; Glimelius, K. Brassica napus lines with rearranged Arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor. Appl. Genet 2003, 106, 1156–1163. [Google Scholar]
- Zubko, M.K.; Zubko, E.I.; Patskovsky, Y.V.; Khvedynich, O.A.; Fisahn, J.; Gleba, Y.Y.; Schieder, O. Novel “homeotic” CMS patterns generated in Nicotiana via cybridiza-tion with Hyoscyamusand Scopolia. J. Exp. Bot 1996, 47, 1101–1110. [Google Scholar]
- Murai, K.; Takumi, S.; Koga, H.; Ogihara, Y. Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. Plant J 2002, 29, 169–181. [Google Scholar]
- Zheng, B.B.; Wu, X.M.; Ge, X.X.; Deng, X.X.; Grosser, J.W.; Guo, W.W. Comparative transcript profiling of a male sterile cybrid pummelo and its fertile type revealed altered gene expression related to flower development. PLoS One 2012, 7, e43758. [Google Scholar]
- Hama, E.; Takumi, S.; Ogihara, Y.; Murai, K. Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 2004, 218, 712–720. [Google Scholar]
- Ma, N.; Liu, C.; Yang, W.; Shen, H. PAP3 regulates stamen but not petal development in Capsicum annuum L. J. Am. Chem. Soc. unpublished work.
- Geng, X.S.; Yang, M.Z.; Huang, X.Q.; Cheng, Z.Q.; FU, J.; Sun, T.; Li, J. Cloning and analyzing of rice blast resistance gene Pi-ta+ allele from Jinghong erect type of common wild rice (Oryza rufipogon Griff) in Yunnan. Hereditas 2008, 30, 109–114. [Google Scholar]
- Zahn, L.M.; Leebens-Mack, J.; DePamphilis, C.W.; Theissen, G. To B or Not to B a flower: The role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered 2005, 96, 225–240. [Google Scholar]
- Munster, T.; Wingen, L.U.; Faigl, W.; Werth, S.; Saedler, H.; Theissen, G. Characterization of three GLOBOSA-like MADS-box genes from maize: Evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 2001, 262, 1–13. [Google Scholar]
- Yu, D.; Kotilainen, M.; Pollanen, E.; Mehto, M.; Elomaa, P.; Helariutta, Y.; Albert, V.A.; Teeri, T.H. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J 1999, 17, 51–62. [Google Scholar]
- Southerton, S.G.; Marshall, H.; Mouradov, A.; Teasdale, R.D. Eucalypt MADS-box genes expressed in developing flowers. Plant Physiol 1998, 118, 365–372. [Google Scholar]
- Skipper, M. Genes from the APETALA3 and PISTILLATA lineages are expressed in developing vascular bundles of the tuberous rhizome, flowering stem and flower Primordia of Eranthis hyemalis. Ann. Bot 2002, 89, 83–88. [Google Scholar]
- Qin, Q.P.; Yin, T.; Chen, J.W.; Xie, M.; Zhang, S.L. APETALA3/DEFICIENS and PISTILLATA/GLOBOSA genes with floral development of plant. Chin. J. Cell Biol 2006, 28, 571–576. [Google Scholar]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development: Anarmchair guide. Nat. Rev. Genet 2005, 6, 688–698. [Google Scholar]
- Carlsson, J.; Lagercrantz, U.; Sundstrom, J.; Teixeira, R.; Wellmer, F.; Meyerowitz, E.M.; Glimelius, K. Microarray analysis reveals altered expression of a large number of nuclear genes in developing cytoplasmic male sterile Brassica napus flowers. Plant J 2007, 49, 452–462. [Google Scholar]
- Yang, J.H.; Qi, X.H.; Zhang, M.F. MADS-box genes are associated with cytoplasmic homeosis in cytoplasmic male-sterile stem mustard as partially mimicked by specifically inhibiting mtETC. Plant Growth Regul 2008, 56, 191–201. [Google Scholar]
- Zhang, J.P.; Gong, Z.H.; Liu, K.K.; Huang, W.; Li, D.W. Interrelation of cytological development period of pepper’s microspore and the morphology of flower organ. J. Northwest A&F Univ 2007, 35, 154–158. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar]
- Zhang, X.; Madi, S.; Borsuk, L.; Nettleton, D.; Elshire, R.J.; Buckner, B.; Janick-Buckner, D.; Beck, J.; Timmermans, M.; Schnable, P.S.; et al. Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet 2007, 3, e101. [Google Scholar]



| Primers | Sequences (5′–3′) |
|---|---|
| F | AGACCTTTTAGGGTTTGAGT |
| R | ACACACTGAATTAAGCAAAA |
| PAP3-F | GGTGGATTAGTTGAGCAGGA |
| PAP3-R | GATGATTTGGTTGAAGGCGT |
| ACTIN-F | AGCACCTCTCAACCCTAA |
| ACTIN-R | GCAAAGCATAACCCTCAT |
| SH-F | GATTTAGGTGACACTATAGAATGCTAGA |
| AAATAGAAAAAAAGTATGGCTC | |
| SH-R | TGTAATACGACTCACTATAGGG |
| ACCTAGACCAAAAGTAGTAATATCA | |
| SL-F | GAAGATCTTCAGAAAATAGAAAAAAAGTATGGCTC |
| SL-R | GGACTAGTCC ACCTAGACCAAAAGTAGTAATATCA |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, X.; Liu, C.; Da, F.; Ma, N.; Shen, H. Expression Pattern of Class B Gene PAP3 in Flower Development of Pepper. Int. J. Mol. Sci. 2013, 14, 24643-24655. https://doi.org/10.3390/ijms141224643
Li X, Liu C, Da F, Ma N, Shen H. Expression Pattern of Class B Gene PAP3 in Flower Development of Pepper. International Journal of Molecular Sciences. 2013; 14(12):24643-24655. https://doi.org/10.3390/ijms141224643
Chicago/Turabian StyleLi, Xin, Chen Liu, Fengjiao Da, Ning Ma, and Huolin Shen. 2013. "Expression Pattern of Class B Gene PAP3 in Flower Development of Pepper" International Journal of Molecular Sciences 14, no. 12: 24643-24655. https://doi.org/10.3390/ijms141224643