CXCL10 Decreases GP73 Expression in Hepatoma Cells at the Early Stage of Hepatitis C Virus (HCV) Infection
Abstract
:1. Introduction
2. Results
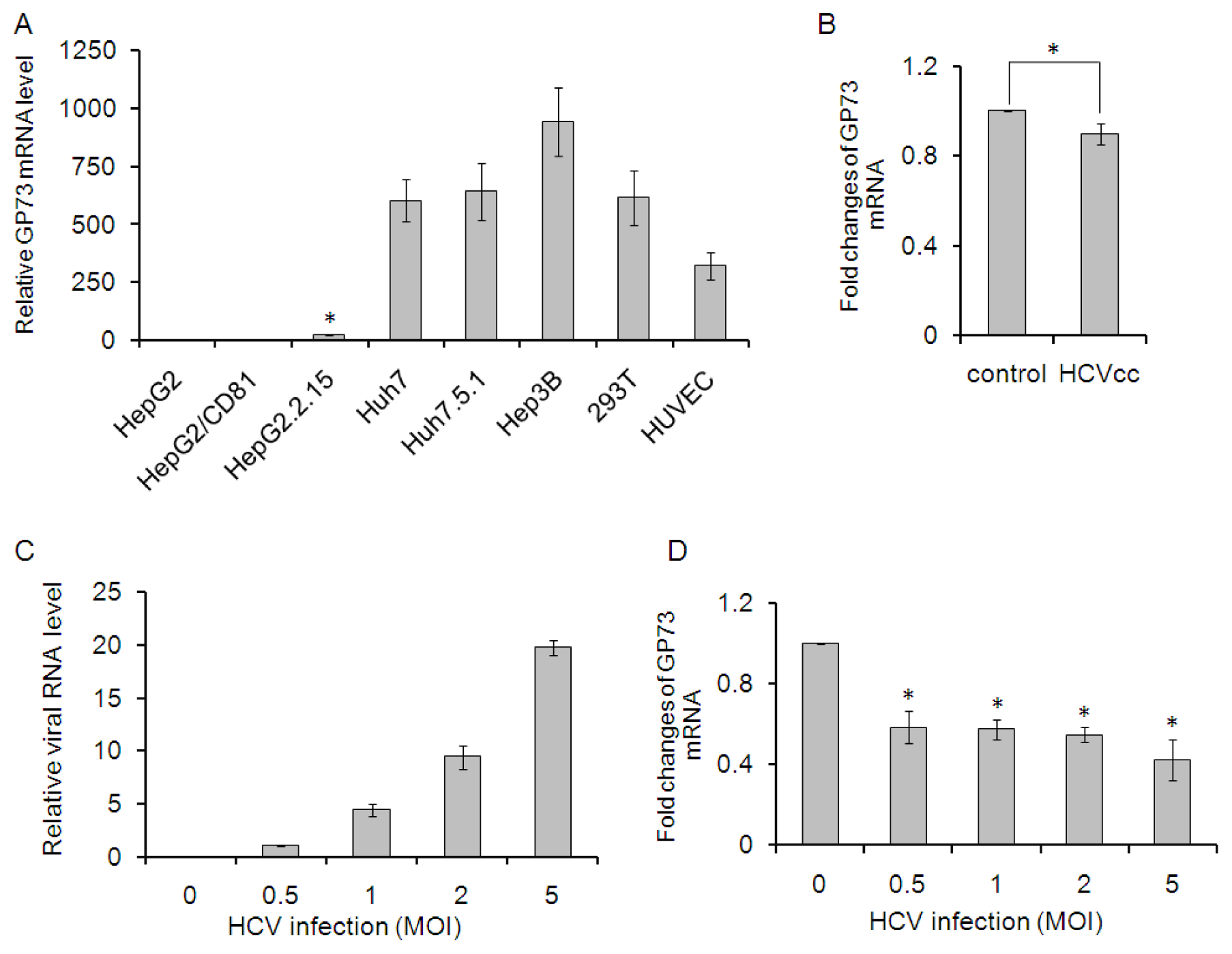
2.1. GP73 mRNA Level Is Down-Regulated in HCV Infected Cells
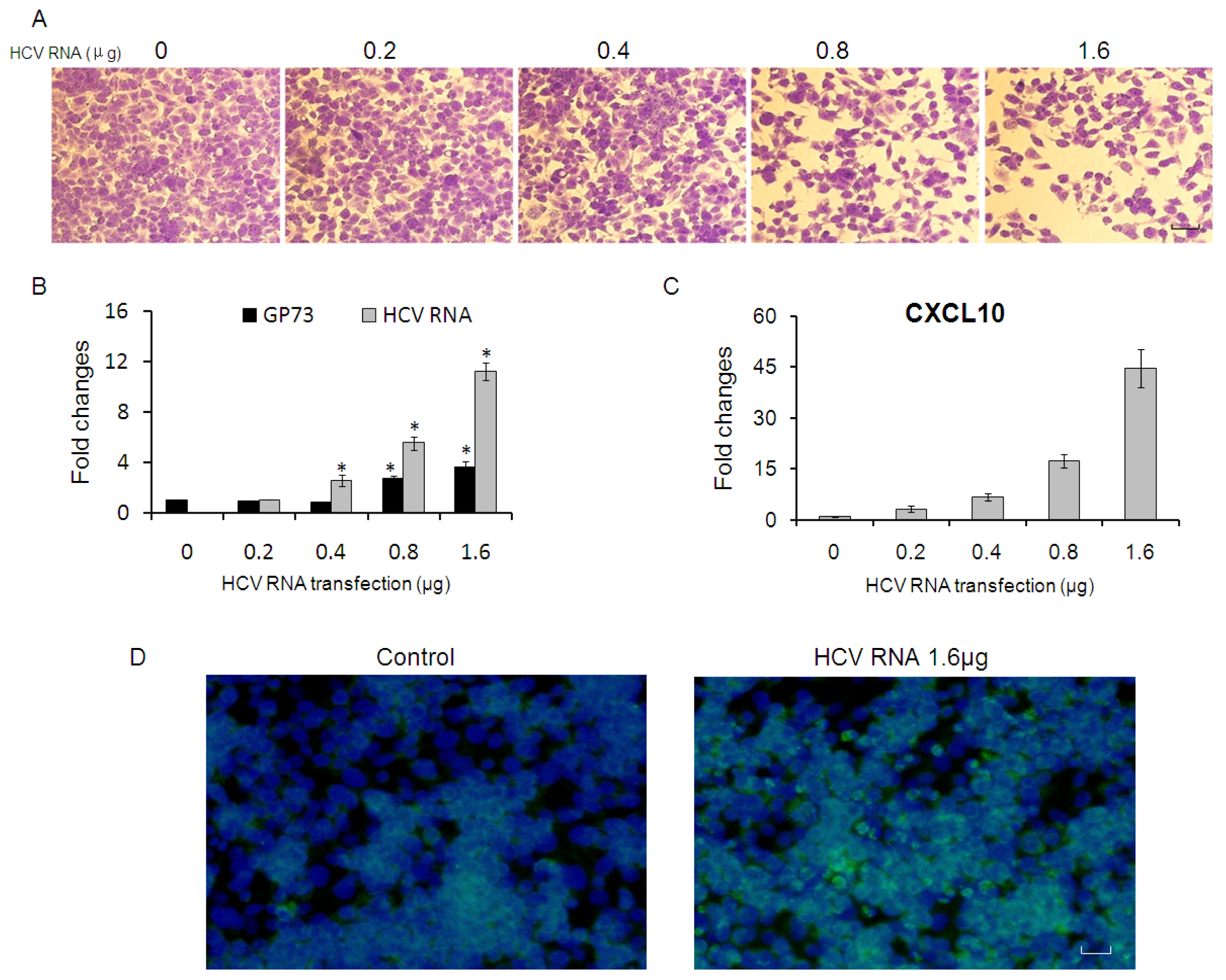
2.2. CXCL10 Is Involved in the Down-Regulation of GP73
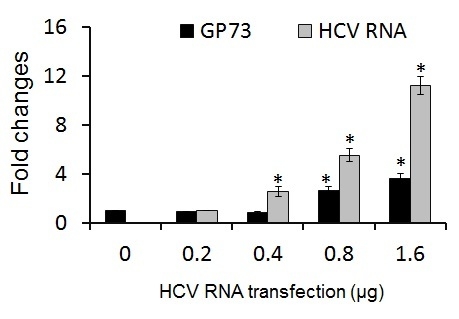
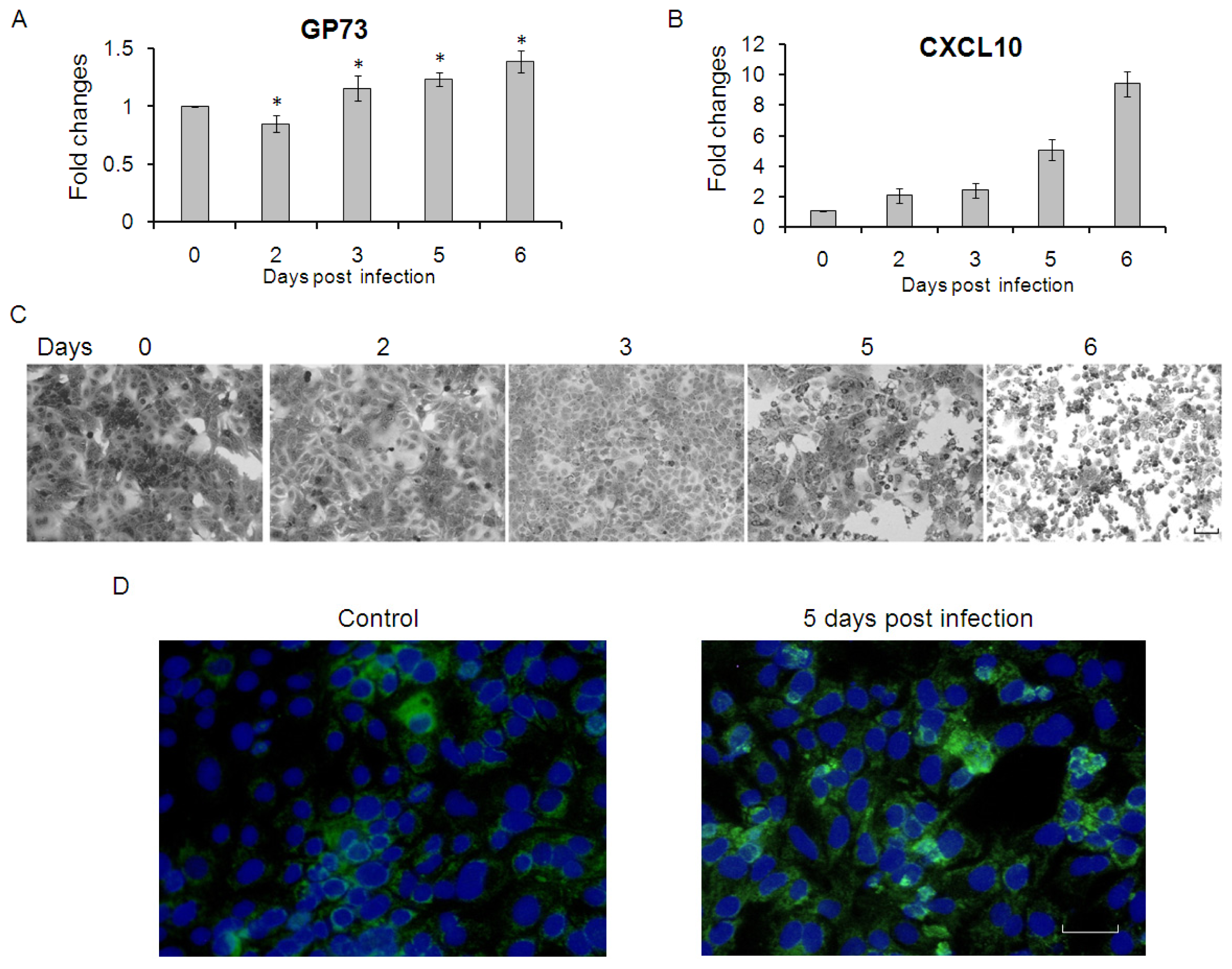
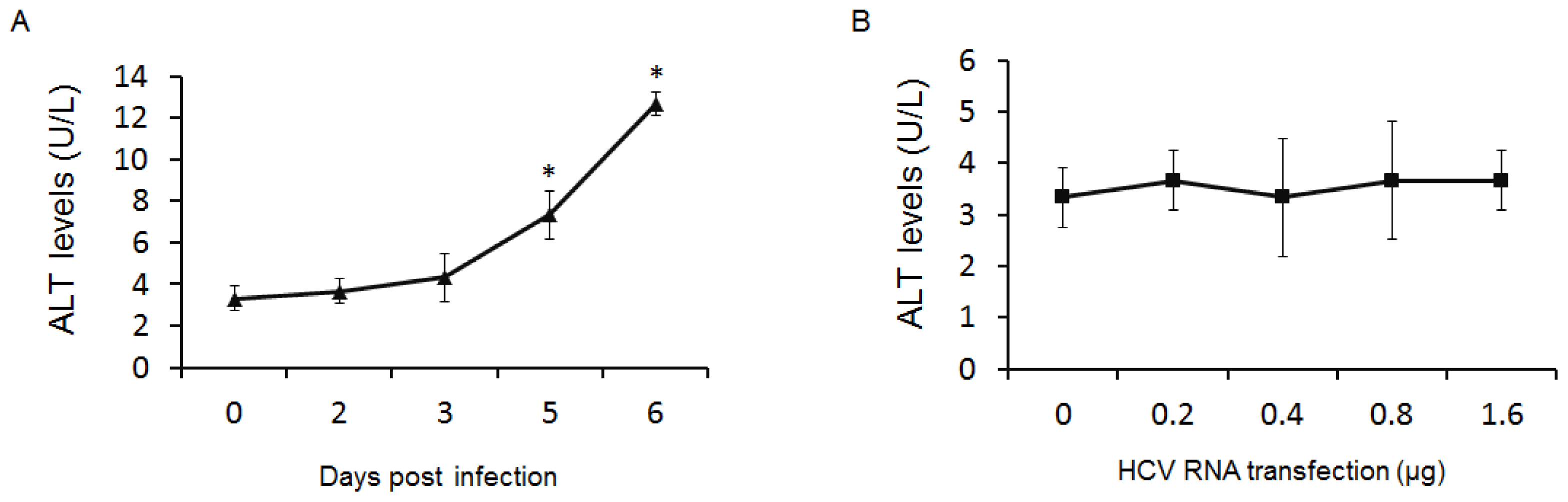
2.3. GP73 Is Up-Regulated in Cells with Cytopathic Effect Induced by HCV Infection
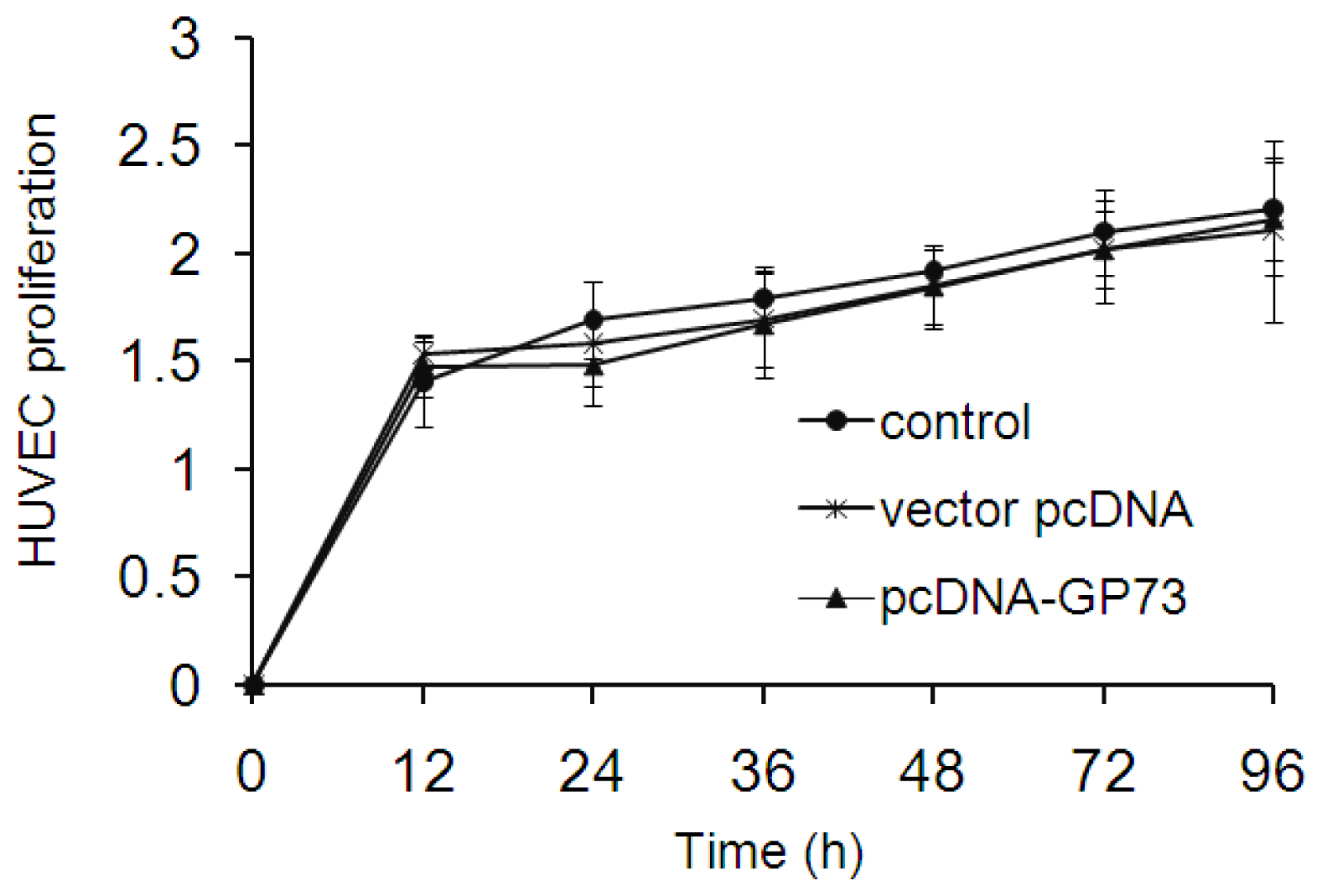
2.4. GP73 Had No Impact on Proliferation of Vascular Endothelial Cells
3. Discussion
4. Materials and Methods
4.1. Cells and Plasmids Construction
4.2. Viral RNA in Vitro Transcriptions
4.3. Real Time PCR
4.4. Cytokine Stimulation
4.5. Immunofluorescence Assay
4.6. Cell Proliferation Assay
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Abbreviations
| GP73 | Golgi protein 73 |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| GOLPH2 | Golgi Phosphoprotein 2 |
| GOLM1 | Golgi membrane protein 1 |
| SNPs | Single-nucleotide polymorphisms |
| AD | Alzheimer’s disease |
| AFP | Alpha-Fetoprotein |
| GPC3 | Glypican-3 |
| DCP | Des-γ-Carboxy Prothrombin |
| CXCL10 | C-X-C motif chemokine 10 |
| CPE | Cytopathic effect |
| HUVEC | Human umbilical vein endothelial cells |
| IFN | Interferon |
| TNF | Tumor necrosis factor |
| IL-6 | Interleukin-6 |
| OSM | oncostatin M |
Conflicts of Interests
References
- Zhou, Y.; Li, L.; Hu, L.; Peng, T. Golgi phosphoprotein 2 (GOLPH2/GP73/GOLM1) interacts with secretory clusterin. Mol. Biol. Rep 2011, 38, 1457–1462. [Google Scholar]
- Kladney, R.D.; Bulla, G.A.; Guo, L.; Mason, A.L.; Tollefson, A.E.; Simon, D.J.; Koutoubi, Z.; Fimmel, C.J. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene 2000, 249, 53–65. [Google Scholar]
- Kladney, R.D.; Tollefson, A.E.; Wold, W.S.; Fimmel, C.J. Upregulation of the Golgi protein GP73 by adenovirus infection requires the E1A CtBP interaction domain. Virology 2002, 301, 236–246. [Google Scholar]
- Marrero, J.A.; Romano, P.R.; Nikolaeva, O.; Steel, L.; Mehta, A.; Fimmel, C.J.; Comunale, M.A.; D’Amelio, A.; Lok, A.S.; Block, T.M. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J. Hepatol 2005, 43, 1007–1012. [Google Scholar]
- Mao, Y.; Yang, H.; Xu, H.; Lu, X.; Sang, X.; Du, S.; Zhao, H.; Chen, W.; Xu, Y.; Chi, T.; et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut 2010, 59, 1687–1693. [Google Scholar]
- Zhang, F.; Gu, Y.; Li, X.; Wang, W.; He, J.; Peng, T. Up-regulated Golgi phosphoprotein 2 (GOLPH2) expression in lung adenocarcinoma tissue. Clin. Biochem 2010, 43, 983–991. [Google Scholar]
- Fritzsche, F.R.; Riener, M.O.; Dietel, M.; Moch, H.; Jung, K.; Kristiansen, G. GOLPH2 expression in renal cell cancer. BMC Urol 2008, 8, 15. [Google Scholar] [Green Version]
- Chen, M.H.; Jan, Y.H.; Chang, P.M.; Chuang, Y.J.; Yeh, Y.C.; Lei, H.J.; Hsiao, M.; Huang, S.F.; Huang, C.Y.; Chau, G.Y. Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Ann. Surg. Oncol 2013, 20, 616–624. [Google Scholar]
- Sun, Y.; Yang, H.; Mao, Y.; Xu, H.; Zhang, J.; Li, G.; Lu, X.; Sang, X.; Zhao, H.; Zhong, S.; et al. Increased Golgi protein 73 expression in hepatocellular carcinoma tissue correlates with tumor aggression but not survival. J. Gastroenterol. Hepatol 2011, 26, 1207–1212. [Google Scholar]
- Chen, L.G.; Wang, H.J.; Yao, H.B.; Guan, T.P.; Wu, F.; He, X.J.; Ma, Y.Y.; Tao, H.Q.; Ye, Z.Y. GP73 is down-regulated in gastric cancer and associated with tumor differentiation. World J. Surg. Oncol 2013, 11, 132. [Google Scholar]
- Yuan, Q.; Chu, C.; Jia, J. Association studies of 19 candidate SNPs with sporadic Alzheimer’s disease in the North Chinese Han population. Neurol. Sci 2012, 33, 1021–1028. [Google Scholar]
- Lin, K.; Tang, M.; Han, H.; Guo, Y.; Lin, Y.; Ma, C. Association between the polymorphisms of CALHM1 and GOLPH2 genes and Alzheimer’s disease. Psychiatr. Genet 2010, 20, 190. [Google Scholar]
- But, D.Y.; Lai, C.L.; Yuen, M.F. Natural history of hepatitis-related hepatocellular carcinoma. World J. Gastroenterol 2008, 14, 1652–1656. [Google Scholar]
- Kladney, R.D.; Cui, X.; Bulla, G.A.; Brunt, E.M.; Fimmel, C.J. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology 2002, 35, 1431–1440. [Google Scholar]
- Iftikhar, R.; Kladney, R.D.; Havlioglu, N.; Schmitt-Gräff, A.; Gusmirovic, I.; Solomon, H.; Luxon, B.A.; Bacon, B.R.; Fimmel, C.J. Disease- and cell-specific expression of GP73 in human liver disease. Am. J. Gastroenterol 2004, 99, 1087–1095. [Google Scholar]
- Flint, M.; von Hahn, T.; Zhang, J.; Farquhar, M.; Jones, C.T.; Balfe, P.; Rice, C.M.; McKeating, J.A. Diverse CD81 proteins support hepatitis C virus infection. J. Virol 2006, 80, 11331–11342. [Google Scholar]
- Zhai, Y.; Shen, X.D.; Gao, F.; Zhao, A.; Freitas, M.C.; Lassman, C.; Luster, A.D.; Busuttil, R.W.; Kupiec-Weglinski, J.W. CXCL10 regulates liver innate immune response against ischemia and reperfusion injury. Hepatology 2008, 47, 207–214. [Google Scholar]
- Wei, H.; Li, B.; Zhang, R.; Hao, X.; Huang, Y.; Qiao, Y.; Hou, J.; Li, X.; Li, X. Serum GP73, a marker for evaluating progression in patients with chronic HBV infections. PLoS One 2013, 8, e53862. [Google Scholar]
- Liang, H.; Block, T.M.; Wang, M.; Nefsky, B.; Long, R.; Hafner, J.; Mehta, A.S.; Marrero, J.; Gish, R.; Norton, P.A. Interleukin-6 and oncostatin M are elevated in liver disease in conjunction with candidate hepatocellular carcinoma biomarker GP73. Cancer Biomark 2012, 11, 161–171. [Google Scholar]
- Tong, Y.; Zhu, Y.; Xia, X.; Liu, Y.; Feng, Y.; Hua, X.; Chen, Z.; Ding, H.; Gao, L.; Wang, Y. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J. Virol 2011, 85, 2793–2802. [Google Scholar]
- Hong, B.; Lee, S.H.; Song, X.T.; Jones, L.; Machida, K.; Huang, X.F.; Chen, S.Y. A super TLR agonist to improve efficacy of dendritic cell vaccine in induction of anti-HCV immunity. PLoS One 2012, 7, e48614. [Google Scholar]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med 2005, 11, 791–796. [Google Scholar]
- Nagpal, M.L.; Davis, J.; Lin, T. Overexpression of CXCL10 in human prostate LNCaP cells activates its receptor (CXCR3) expression and inhibits cell proliferation. Biochim. Biophys. Acta 2006, 1762, 811–818. [Google Scholar]

| Gene | Primer sequences (5′-3′) |
|---|---|
| GP73 | Forward:CATAGAAGCTTCTCCAGACACGGATCATGGAGCTGGAAGGC |
| Reverse:GAGAGGGATCCTCAGAGTGTATGATTCCGCTTTTCACGCTG | |
| CXCL10 | Forward:CATAGAAGCTTCTCGAGATGCTCCTGGCTGTTTTGTACTGCCTGCTGTGGAGTTTCCAGACCTCCGCTGGCCATTTCCCTAGAATGAATCAAACTGCCATTCTG |
| Reverse:GAGAGGGATCCAACATAGCACCTCAGTAGAGC |
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| HCV | CTTCACGCAGAAAGCGTCTA | CAAGCACCCTATCAGGCAGT |
| GP73 | TTGGTAACAGCAAGTCCCAGACA | ACCACCTGGATCTCATTGGTTTC |
| CXCL10 | AGGAACCTCCAGTCTCAGCA | CAAAATTGGCTTGCAGGAAT |
| GAPDH | TGGGCTACACTGAGCACCAG | AAGTGGTCGTTGAGGGCAAT |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, Y.; Zou, Z.; Zhu, B.; Hu, Z.; Zeng, P. CXCL10 Decreases GP73 Expression in Hepatoma Cells at the Early Stage of Hepatitis C Virus (HCV) Infection. Int. J. Mol. Sci. 2013, 14, 24230-24241. https://doi.org/10.3390/ijms141224230
Liu Y, Zou Z, Zhu B, Hu Z, Zeng P. CXCL10 Decreases GP73 Expression in Hepatoma Cells at the Early Stage of Hepatitis C Virus (HCV) Infection. International Journal of Molecular Sciences. 2013; 14(12):24230-24241. https://doi.org/10.3390/ijms141224230
Chicago/Turabian StyleLiu, Yuan, Ziying Zou, Bing Zhu, Zonghai Hu, and Ping Zeng. 2013. "CXCL10 Decreases GP73 Expression in Hepatoma Cells at the Early Stage of Hepatitis C Virus (HCV) Infection" International Journal of Molecular Sciences 14, no. 12: 24230-24241. https://doi.org/10.3390/ijms141224230