Design, Synthesis, Antifungal Activities and 3D-QSAR of New N,N'-Diacylhydrazines Containing 2,4-Dichlorophenoxy Moiety
Abstract
:1. Introduction
2. Results and Discussion
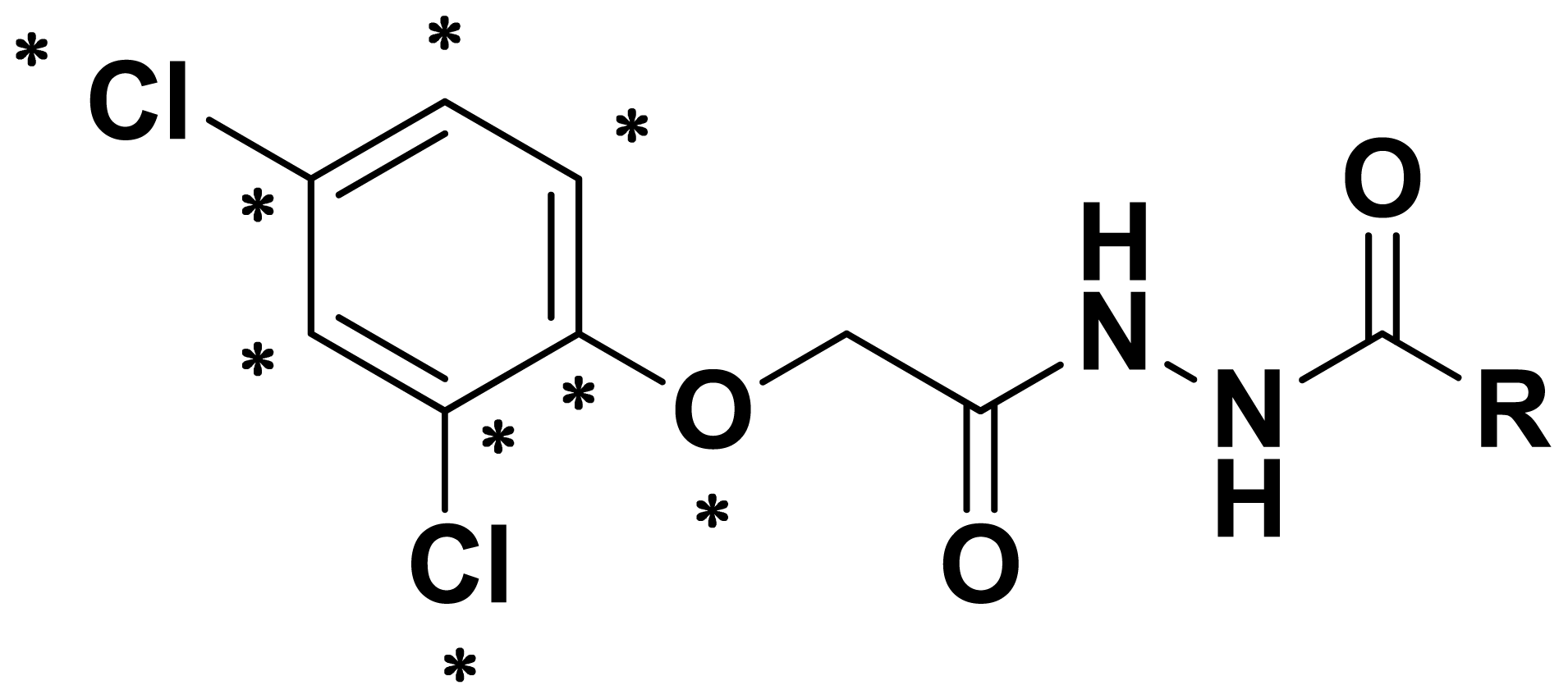
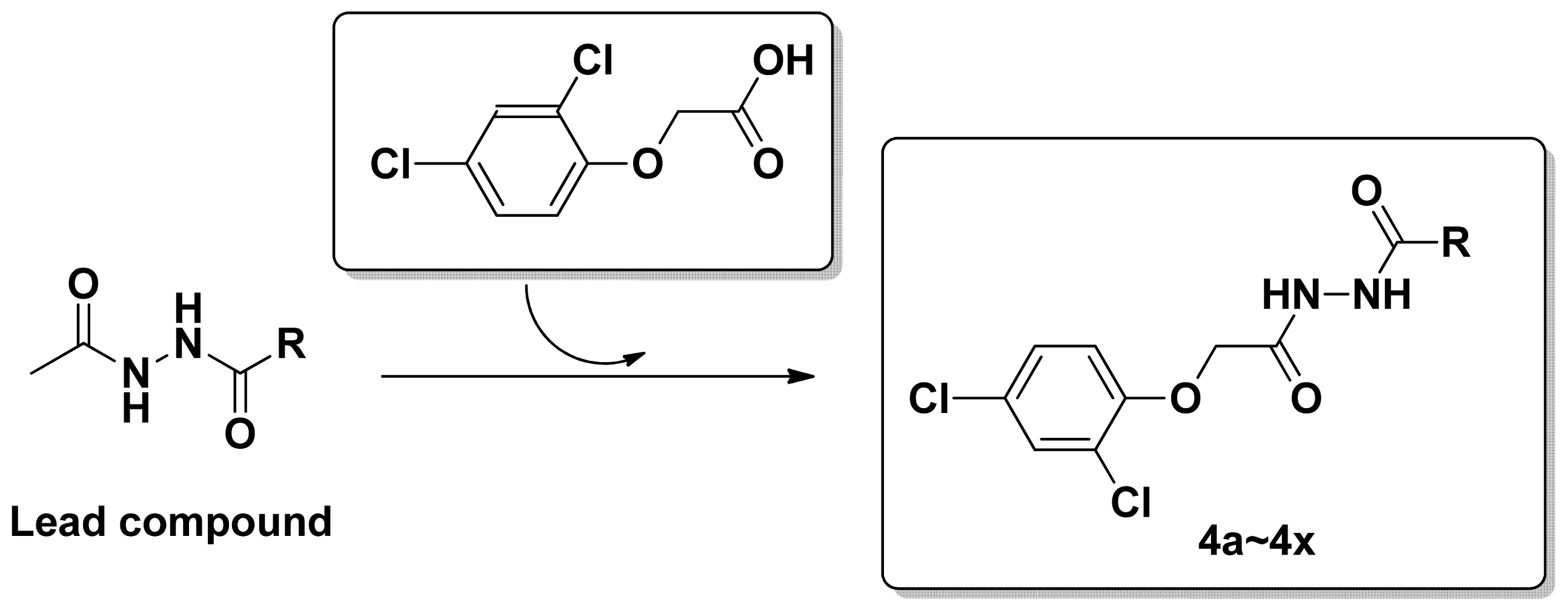
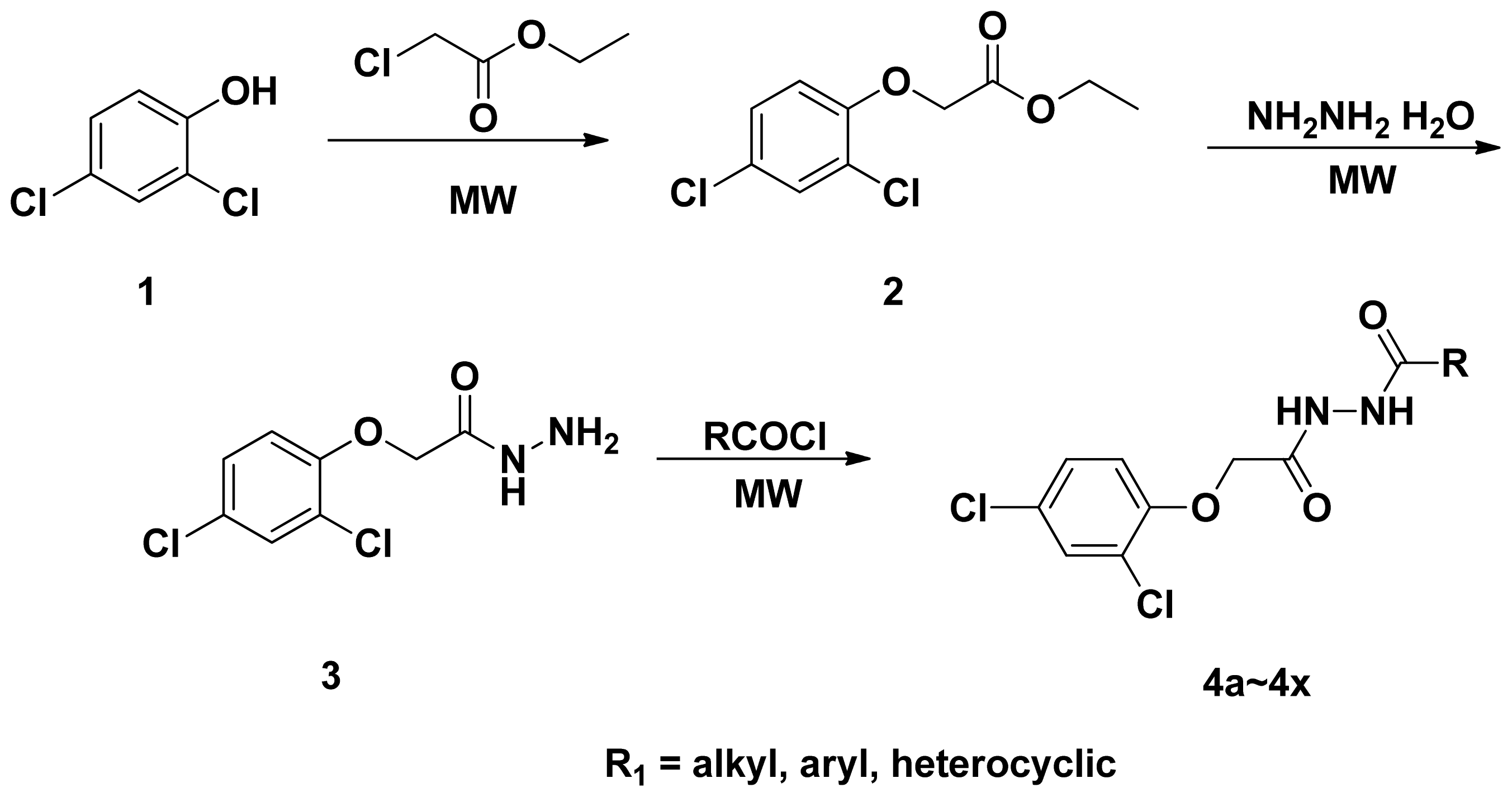
2.1. Synthesis and Spectrum
2.2. Antifungal Activities
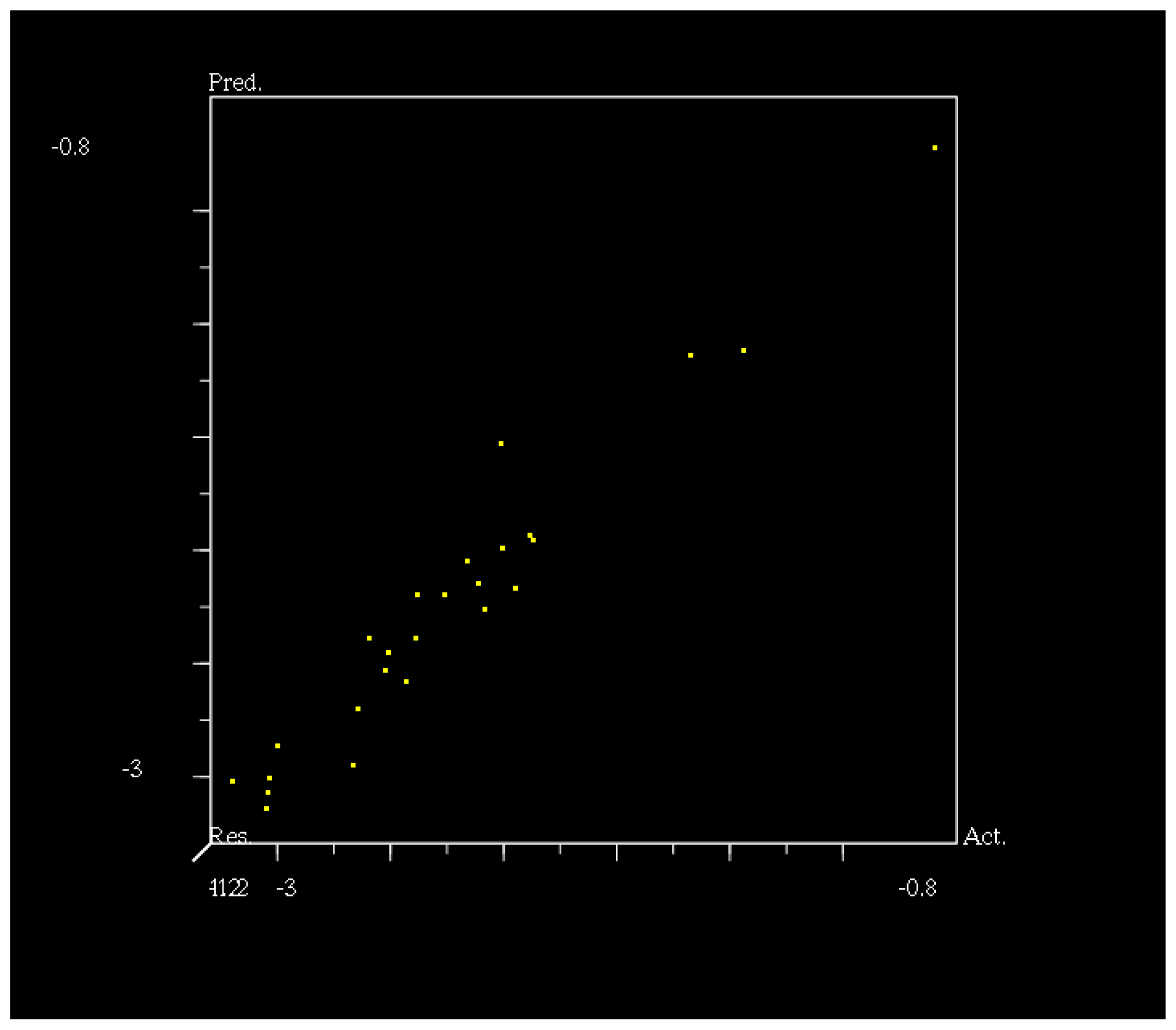
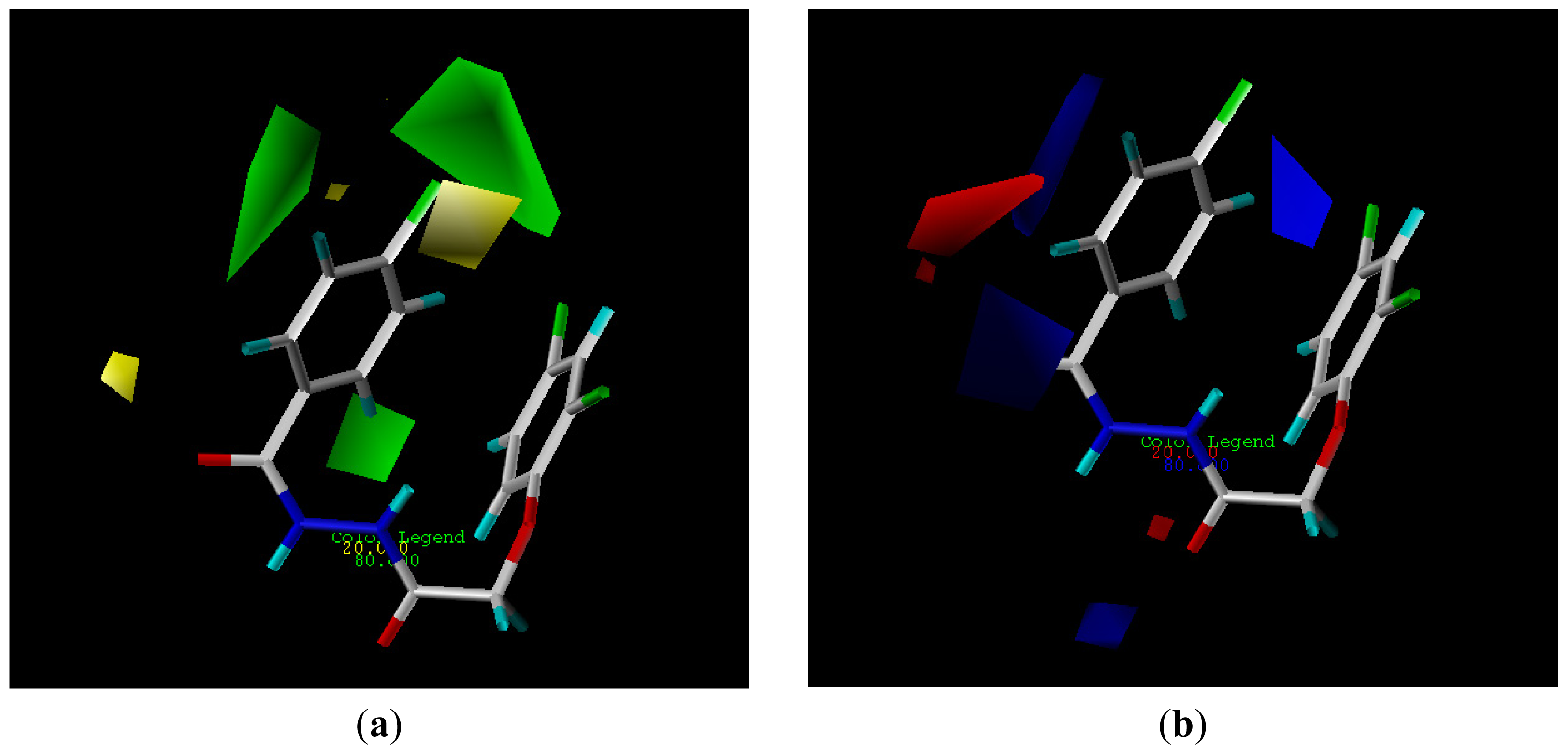
2.3. CoMFA Analysis
3. Experimental Section
3.1. Instruments
3.2. General Procedure
N′-[2-(2,4-Dichlorophenoxy)acetyl]cyclopropanecarbohydrazide 4a
N′-[2-(2,4-Dichlorophenoxy)acetyl]benzohydrazide 4b
N′-[2-(2,4-Dichlorophenoxy)acetyl]-4-nitrobenzohydrazide 4c
4-Chloro-N′-[2-(2,4-dichlorophenoxy)acetyl]benzohydrazide 4d
N′-[2-(2,4-Dichlorophenoxy)acetyl]-3-methylbenzohydrazide 4e
3-Chloro-N′-[2-(2,4-dichlorophenoxy)acetyl]benzohydrazide 4f
N′-[2-(2,4-Dichlorophenoxy)acetyl]-2-fluorobenzohydrazide 4g
2-Chloro-N′-[2-(2,4-dichlorophenoxy)acetyl]benzohydrazide 4h
2,4-Dichloro-N′-[2-(2,4-dichlorophenoxy)acetyl]benzohydrazide 4i
N′-[2-(2,4-Dichlorophenoxy)acetyl]-2-methoxybenzohydrazide 4j
N′-[2-(2,4-Dichlorophenoxy)acetyl]-4-methoxybenzohydrazide 4k
N′-[2-(2,4-Dichlorophenoxy)acetyl]-4-iodobenzohydrazide 4l
N′-[2-(2,4-Dichlorophenoxy)acetyl]-5-methylisoxazole-4-carbohydrazide 4m
1-Cyano-N′-[2-(2,4-dichlorophenoxy)acetyl]cyclopropanecarbohydrazide 4n
N′-[2-(2,4-Dichlorophenoxy)acetyl] butyrohydrazide 4o
N′-[2-(2,4-Dichlorophenoxy)acetyl]isobutyrohydrazide 4p
N′-[2-(2,4-Dichlorophenoxy)acetyl]butyrohydrazide 4q
N′-[2-(2,4-Dichlorophenoxy)acetyl]pentanehydrazide 4r
2-(2,4-Dichlorophenoxy)-N′-[2-(2,4-dichlorophenoxy)acetyl]acetohydrazide 4s
2-(2,4-Dichlorophenoxy)-N′-[2-(2,4-dichlorophenoxy)acetyl]propanehydrazide 4t
N′-[2-(2,4-Dichlorophenoxy)acetyl]furan-3-carbohydrazide 4u
(2E,4Z)-N′-[2-(2,4-Dichlorophenoxy)acetyl]hexa-2,4-dienehydrazide 4v
N′-[2-(2,4-Dichlorophenoxy)acetyl]nicotinohydrazide 4w
N′-[2-(2,4-Dichlorophenoxy)acetyl]isonicotinohydrazide 4x
3.3. 3D-QSAR Analysis
3.4. Antifungal Activities Assay
4. Conclusions

| No. | Corynespora cassiicola | Cladosporium cucumerinum | Sphaerotheca fuligenea | Sclerotinia sclerotiorum | Colletotrichum orbiculare |
|---|---|---|---|---|---|
| 4a | 10.00 | 10.00 | 59.09 | 13.82 | 89.79 |
| 4b | 49.00 | 20.00 | 100.00 | 15.55 | 72.52 |
| 4c | 37.00 | 37.00 | 34.54 | nd | 72.77 |
| 4d | 39.00 | 59.00 | 18.18 | 49.49 | 98.75 |
| 4e | 42.00 | 4.00 | 42.70 | 42.23 | 68.70 |
| 4f | 38.00 | nd | 77.60 | 51.17 | 46.48 |
| 4g | 38.00 | 19.00 | 35.71 | 31.85 | 66.94 |
| 4h | 38.00 | 71.00 | 67.27 | 16.14 | 19.87 |
| 4i | nd | nd | nd | −4.24 | 25.85 |
| 4j | 42.00 | 7.00 | 12.34 | 13.97 | 43.18 |
| 4k | 41.00 | 49.00 | 30.30 | 31.06 | 63.46 |
| 4l | 35.00 | 13.00 | 82.85 | 6.51 | 51.21 |
| 4m | 38.00 | 4.00 | 88.31 | 22.18 | 64.47 |
| 4n | 27.00 | 6.00 | 57.14 | 55.13 | 23.92 |
| 4o | 25.00 | 6.00 | 38.22 | 33.34 | 50.82 |
| 4p | 85.00 | 1.00 | 62.60 | 16.51 | 39.28 |
| 4q | 87.00 | 53.00 | −5.19 | 25.19 | 93.11 |
| 4r | 25.00 | 5.00 | 15.58 | 7.66 | 71.91 |
| 4s | 54.00 | 4.00 | 79.16 | 30.39 | 28.85 |
| 4t | 45.00 | 44.00 | 45.45 | nd | 46.63 |
| 4u | 10.00 | 55.00 | 41.23 | 28.12 | 67.45 |
| 4v | 64.00 | 15.00 | 6.49 | 1.60 | 24.88 |
| 4w | 61.00 | 24.00 | 47.06 | 23.33 | 44.92 |
| 4x | 48.00 | 47.00 | 8.16 | 29.45 | 63.76 |
| chlorothalonil | 69.93 | 56.27 | 12.12 | 53.40 | 39.41 |
| Compound | Fungi | EC50 |
|---|---|---|
| 4b | S. fuligenea | 11.2287 |
| flusilazole | S. fuligenea | 0.8923 |
| No. | R | D | D″ | Residue |
|---|---|---|---|---|
| 4a | cycloprane | −1.53744 | −1.6016 | 0.06416 |
| 4b | phenyl | −2.10898 | −2.2012 | 0.09222 |
| 4c | p-nitro phenyl | −2.15762 | −2.1771 | 0.01948 |
| 4d* | p-chloro phenyl | −0.6748 | −0.6752 | 0.0004 |
| 4e | p-fluoro phenyl | −2.21145 | −2.2910 | 0.07955 |
| 4f | m-methyl phenyl | −2.60927 | −2.5781 | −0.03117 |
| 4g# | m-chloro phenyl | −2.26605 | −2.3169 | 0.05085 |
| 4h | o-F phenyl | −3.15846 | −3.0991 | −0.05936 |
| 4i | o-chloro phenyl | −3.03008 | −2.9571 | −0.07298 |
| 4j | 2,4-dichloro phenyl | −2.72994 | −2.80212 | 0.07218 |
| 4k | o-methoxyl phenyl | −2.32753 | −2.4336 | 0.10607 |
| 4l | p-OMe Ph | −2.54624 | −2.6018 | 0.05556 |
| 4m | p-iodo phenyl | −2.40876 | −2.5266 | 0.11784 |
| 4n | isoxazoyl | −3.03926 | −3.1169 | 0.07764 |
| 4o | 1-cycan-cyclopropyl | −2.50183 | −2.4923 | −0.00953 |
| 4p | propyl | −2.67369 | −2.5563 | −0.11739 |
| 4q | Iso-propyl | −1.35375 | −1.3239 | −0.02985 |
| 4r | Butyl | −2.0958 | −2.1276 | 0.0318 |
| 4s | 2,4-dichlorophenoxymethyl | −3.03359 | −3.1659 | 0.13231 |
| 4t# | [2-(2,4-dichlorophenoxy)acetyl]propyl | −2.71389 | −2.6361 | −0.07779 |
| 4u | furan | −2.20095 | −2.3172 | 0.11625 |
| 4v | (2E,4Z)-hex | −2.99734 | −3.1581 | 0.16076 |
| 4w | 3-pyridine | −2.62024 | −2.7768 | 0.15656 |
| 4x# | 4-pyridine | −2.28632 | −2.4019 | 0.11558 |
Acknowledgments
Conflicts of Interest
References
- Kim, B.Y.; Willbold, S.; Kulik, A.; Helaly, S.E.; Zinecker, H.; Wiese, J.; Imhoff, J.F.; Goodfellow, M.; Sussmuth, R.D.; Fiedler, H.P. Elaiomycins B and C, novel alkylhydrazides produced by Streptomyces sp. BK 190. J. Antibiot 2011, 64, 595–597. [Google Scholar]
- Shoeb, M.; MacManus, S.M.; Jaspars, M.; Trevidu, J.; Nahar, L.; Kong-Thoo-Lin, P.; Sarker, S.D. Montamine, a unique dimeric indole alkaloid, from the seeds of Centaurea montana (Asteraceae), and its in vitro cytotoxic activity against the CaCo2 colon cancer cells. Tetrahedron 2006, 62, 11172–11177. [Google Scholar]
- Zheng, J.; Liu, C.; Sawaya, M.R.; Vadla, B.; Khan, S.; Woods, R.J.; Eisenberg, D.; Goux, W.J.; Nowick, J.S. Macrocyclic β-sheet peptides that inhibit the aggregation of a tau-protein-derived hexapeptide. J. Am. Chem. Soc 2011, 133, 3144–3157. [Google Scholar]
- Zhao, Q.Q.; Ou, X.M.; Huang, Z.Q.; Bi, F.C.; Huang, R.Q.; Wang, Q.M. Synthesis and insecticidal activities of novel N-sulfenyl-N′-tert-butyl-N,N′-diacylhydrazines. 3. N-(alkyldithio), N-(aminothio), and N,N-dithio derivatives. J. Agric. Food Chem 2008, 56, 10799–10804. [Google Scholar]
- Zhao, H.; Neamati, N.; Sunder, S.; Hong, H.X.; Wang, S.M.; Milne, G.W.A.; Pommier, Y.; Burke, T.R. Hydrazide-containing inhibitors of HIV-1 integrase. J. Med. Chem 1997, 40, 937–941. [Google Scholar]
- Liu, X.H.; Shi, Y.X.; Ma, Y.; He, G.R.; Dong, W.L.; Zhang, C.Y.; Wang, B.L.; Wang, S.H.; Li, B.J.; Li, Z.M. Synthesis of some N,N′-diacylhydrazine derivatives with radical-scavenging and antifungal activity. Chem. Biol. Drug Des 2009, 73, 320–327. [Google Scholar]
- Li, W.; Gao, L.M.; Zhang, Y.M.; Wei, T.B. Synthesis and biological activity of 1-[2-(1-phenyl-1H-tetrazole-5-yl)-thio] acetylhydrazine derivatives (in Chinese). Huaxue Shiji 2008, 30, 89–91. [Google Scholar]
- Liu, X.H.; Pan, L.; Ma, Y.; Weng, J.Q.; Tan, C.X.; Li, Y.H.; Shi, Y.X.; Li, B.J.; Li, Z.M.; Zhang, Y.G. Design, synthesis, biological activities, and 3D-QSAR of new N,N′-diacylhydrazines containing 2-(2,4-dichlorophenoxy)propane moiety. Chem. Biol. Drug Des 2011, 78, 689–694. [Google Scholar]
- Liu, H.; Wang, H.Q.; Liu, Z.J. Synthesis and herbicidal activity of novel pyrazolo[3,4-d]pyrimidin-4-one derivatives containing aryloxyphenoxypropionate moieties. Bioorg. Med. Chem. Lett 2007, 17, 2203–2209. [Google Scholar]
- Han, L.; Gao, J.R.; Li, Z.M.; Zhang, Y.; Guo, W.M. Synthesis of new plant growth regulator: N-(Fatty acid) O-aryloxyacetyl ethanolamine. Bioorg. Med. Chem. Lett 2007, 17, 3231–3234. [Google Scholar]
- Zhang, H.; Zan, J.H.; Yu, G.Y.; Jiang, M.; Liu, P.X. A combination of 3D-QSAR, molecular docking and molecular dynamics simulation studies of benzimidazole-quinolinone derivatives as iNOS inhibitors. Int. J. Mol. Sci 2012, 13, 11210–11227. [Google Scholar]
- Lu, X.Y.; Lv, M.; Huang, K.; Ding, K.; You, Q.D. Pharmacophore and molecular docking guided 3D-QSAR study of bacterial enoyl-ACP reductase (FabI) inhibitors. Int. J. Mol. Sci 2012, 13, 6620–6638. [Google Scholar]
- Ouyang, L.; He, G.; Huang, W.; Song, X.R.; Wu, F.B.; Xiang, M.L. Combined structure-based pharmacophore and 3D-QSAR studies on phenylalanine series compounds as TPH1 inhibitors. Int. J. Mol. Sci 2012, 13, 5348–5363. [Google Scholar]
- Hansch, C.; Fujita, T. ρ-σ-π analysis. Method for correlation of biological activity chemical structure. J. Am. Chem. Soc 1964, 86, 1616–1626. [Google Scholar]
- Free, S.M.; Wilson, J.W. A mathematical contribution to structure-activity studies. J. Med. Chem 1964, 7, 395–399. [Google Scholar]
- Putz, M.V.; Lazea, M.; Putz, A.M.; Duda-Seiman, C. Introducing catastrophe-QSAR. Application on modeling molecular mechanisms of pyridinone derivative-type HIV non-nucleoside reverse transcriptase inhibitors. Int. J. Mol. Sci 2011, 12, 9533–9569. [Google Scholar]
- Putz, M.V.; Ionascu, C.; Putz, A.M.; Ostafe, V. Alert-QSAR. Implications for electrophilic theory of chemical carcinogenesis. Int. J. Mol. Sci 2011, 12, 5098–5134. [Google Scholar]
- Putz, M.V.; Lazea, M.; Sandjo, L.P. Quantitative structure inter-activity relationship (QSInAR). Cytotoxicity study of some hemisynthetic and isolated natural steroids and precursors on human fibrosarcoma cells HT1080. Molecules 2011, 16, 6603–6620. [Google Scholar]
- Putz, M.V.; Putz, A.M.; Lazea, M.; Ienciu, L.; Chiriac, A. Quantum-SAR extension of the spectral-SAR algorithm. Application to polyphenolic anticancer bioactivity. Int. J. Mol. Sci 2009, 10, 1193–1214. [Google Scholar]
- De Souza, S.D.; de Souza, A.M.T.; de Sousa, A.C.C.; Sodero, A.C.R.; Cabral, L.M.; Albuquerque, M.G.; Castro, H.C.; Rodrigues, C.R. Hologram QSAR models of 4-[(diethylamino)methyl]-phenol inhibitors of acetyl/butyrylcholinesterase enzymes as potential anti-alzheimer agents. Molecules 2012, 17, 9529–9539. [Google Scholar]
- Putz, M.V. Residual-QSAR. Implications for genotoxic carcinogenesis. Chem. Cent. J 2011, 5, 1–11. [Google Scholar]
- Putz, M.V.; Dudaş, N.A. Determining chemical reactivity driving biological activity from SMILES transformations: The bonding mechanism of anti-HIV pyrimidines. Molecules 2013, 18, 9061–9116. [Google Scholar]
- Putz, M.V.; Dudaş, N.A. Variational principles for mechanistic quantitative structure–activity relationship (QSAR) studies: Application on uracil derivatives’ anti-HIV action. Struct. Chem 2013. [Google Scholar] [CrossRef]
- Putz, M.V.; Putz, A.M.; Barou, R. Spectral-SAR realization of OECD-QSAR principles. Int. J. Chem. Model 2011, 3, 173–190. [Google Scholar]
- Liu, X.H.; Weng, J.Q.; Tan, C.X. Microwave synthesis and biological activity of hydrazone derivatives containing 1,2,3-thiadiazole. Asian J. Chem 2011, 23, 4064–4066. [Google Scholar]
- Liu, X.H.; Chen, P.Q.; Wang, B.L.; Li, Y.H.; Wang, S.H.; Li, Z.M. Synthesis, bioactivity, theoretical and molecular docking study of 1-cyano-N-substituted-cyclopropanecarboxamide as ketol-acid reductoisomerase inhibitor. Bioorg. Med. Chem. Lett 2007, 17, 3784–3788. [Google Scholar]
- Xue, Y.L.; Liu, X.H.; Zhang, Y.G. Synthesis, crystal structure and biological activity of 1-cyano-N-phenylcyclopropanecarboxamide. Asian J. Chem 2012, 24, 1571–1574. [Google Scholar]
- Xue, Y.L.; Zhang, Y.G.; Liu, X.H. Synthesis, crystal structure and biological activity of 1-Cyano-N-(4-bromophenyl)cyclopropanecarboxamide. Asian J. Chem 2012, 24, 3016–3018. [Google Scholar]
- Wu, R.; Zhu, C.; Du, X.J.; Xiong, L.X.; Yu, S.J.; Liu, X.H.; Li, Z.M.; Zhao, W.G. Synthesis, crystal structure and larvicidal activity of novel diamide derivatives against. Culex pipiens. Chem. Cent. J 2012, 6, 1–5. [Google Scholar]
- Weng, J.Q.; Wang, L.; Liu, X.H. Synthesis, crystal structure and herbicidal activity of a 1, 2, 4-triazol-5 (4H)-one derivative. J. Chem. Soc. Pak 2012, 34, 1248–1252. [Google Scholar]
- Xue, Y.L.; Zhang, Y.G.; Liu, X.H. Synthesis, crystal structure and biological activity of 1-cyano-N-(2,4-dichlorophenyl)cyclopropanecarboxamide. Asian J. Chem 2012, 24, 5087–5089. [Google Scholar]
- Cui, C.; Wang, Z.P.; Du, X.J.; Wang, L.Z.; Yu, S.J.; Liu, X.H.; Li, Z.M.; Zhao, W.G. Synthesis and antiviral activity of hydrogenated ferulic acid derivatives. J. Chem 2013, 2013, 269434:1–269434:5. [Google Scholar]
- Sun, N.B.; Liu, X.H.; Weng, J.Q.; Tan, C.X. An unexpected product N-{3-[(2-fluorobenzyl)thio]-5-methyl-4H-1,2,4-triazol-4-yl}acetimidamide: Synthesis and structure analysis. J. Chem. Soc. Pak 2013, 35, 499–502. [Google Scholar]
- Huang, Z.Q.; Zhao, Q.Q.; Huang, R.Q.; Wang, Q.M. Design and synthesis of novel N′-tert-butyl-N′-substitutedbenzoyl-N-[dihydrobenzofuran(chroman)]carbohydrazide derivatives as potential insect growth regulators. Lett. Org. Chem 2009, 6, 29–36. [Google Scholar]
- Zhao, Q.Q.; Shang, J.; Huang, Z.Q.; Wang, K.Y.; Bi, F.; Huang, R.Q.; Wang, Q.M. Synthesis and insecticidal activities of novel N-Sulfenyl-N′-tert-butyl-N,N′-diacylhydrazines. 2. N-Substituted phenoxysulfenate derivatives. J. Agr. Food Chem 2008, 56, 5254–5259. [Google Scholar]
- SYBYL, version 6.91; Tripos Inc: St. Louis, MO, USA, 2004.
- Liu, X.H.; Shi, Y.X.; Ma, Y.; Zhang, C.Y.; Dong, W.L.; Pan, L.; Wang, B.L.; Li, B.J.; Li, Z.M. Synthesis, antifungal activities and 3D-QSAR study of N-(5-substituted-1,3,4-thiadiazol-2-yl) cyclopropanecarboxamides. Eur. J. Med. Chem 2009, 44, 2782–2786. [Google Scholar]
- Tan, C.X.; Shi, Y.X.; Weng, J.Q.; Liu, X.H.; Li, B.J.; Zhao, W.G. Synthesis and antifungal activity of 1,2,4-triazole derivatives containing cyclopropane moiety. Lett. Drug Des. Discov 2012, 9, 431–435. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sun, N.-B.; Shi, Y.-X.; Liu, X.-H.; Ma, Y.; Tan, C.-X.; Weng, J.-Q.; Jin, J.-Z.; Li, B.-J. Design, Synthesis, Antifungal Activities and 3D-QSAR of New N,N'-Diacylhydrazines Containing 2,4-Dichlorophenoxy Moiety. Int. J. Mol. Sci. 2013, 14, 21741-21756. https://doi.org/10.3390/ijms141121741
Sun N-B, Shi Y-X, Liu X-H, Ma Y, Tan C-X, Weng J-Q, Jin J-Z, Li B-J. Design, Synthesis, Antifungal Activities and 3D-QSAR of New N,N'-Diacylhydrazines Containing 2,4-Dichlorophenoxy Moiety. International Journal of Molecular Sciences. 2013; 14(11):21741-21756. https://doi.org/10.3390/ijms141121741
Chicago/Turabian StyleSun, Na-Bo, Yan-Xia Shi, Xing-Hai Liu, Yi Ma, Cheng-Xia Tan, Jian-Quan Weng, Jian-Zhong Jin, and Bao-Ju Li. 2013. "Design, Synthesis, Antifungal Activities and 3D-QSAR of New N,N'-Diacylhydrazines Containing 2,4-Dichlorophenoxy Moiety" International Journal of Molecular Sciences 14, no. 11: 21741-21756. https://doi.org/10.3390/ijms141121741





