OsPOP5, A Prolyl Oligopeptidase Family Gene from Rice Confers Abiotic Stress Tolerance in Escherichia coli
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
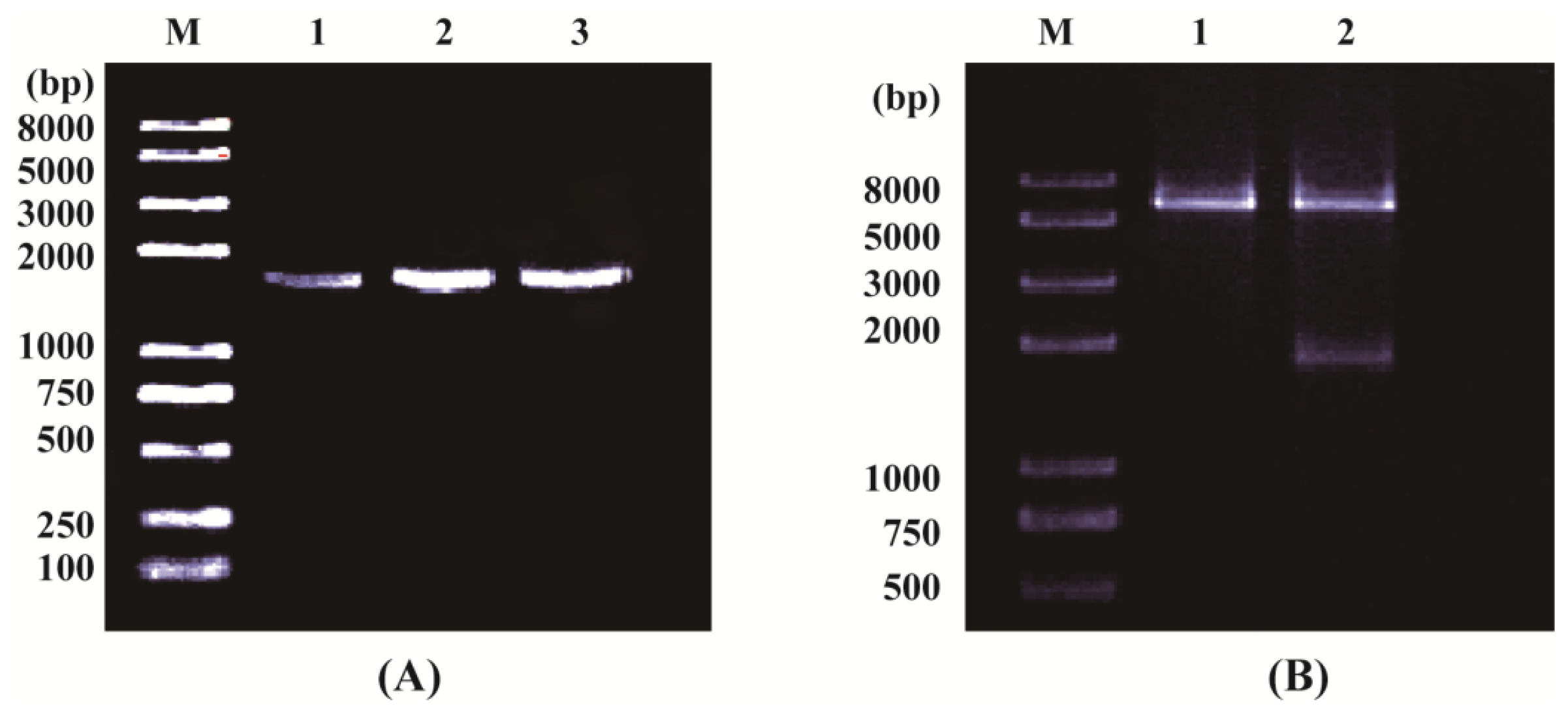
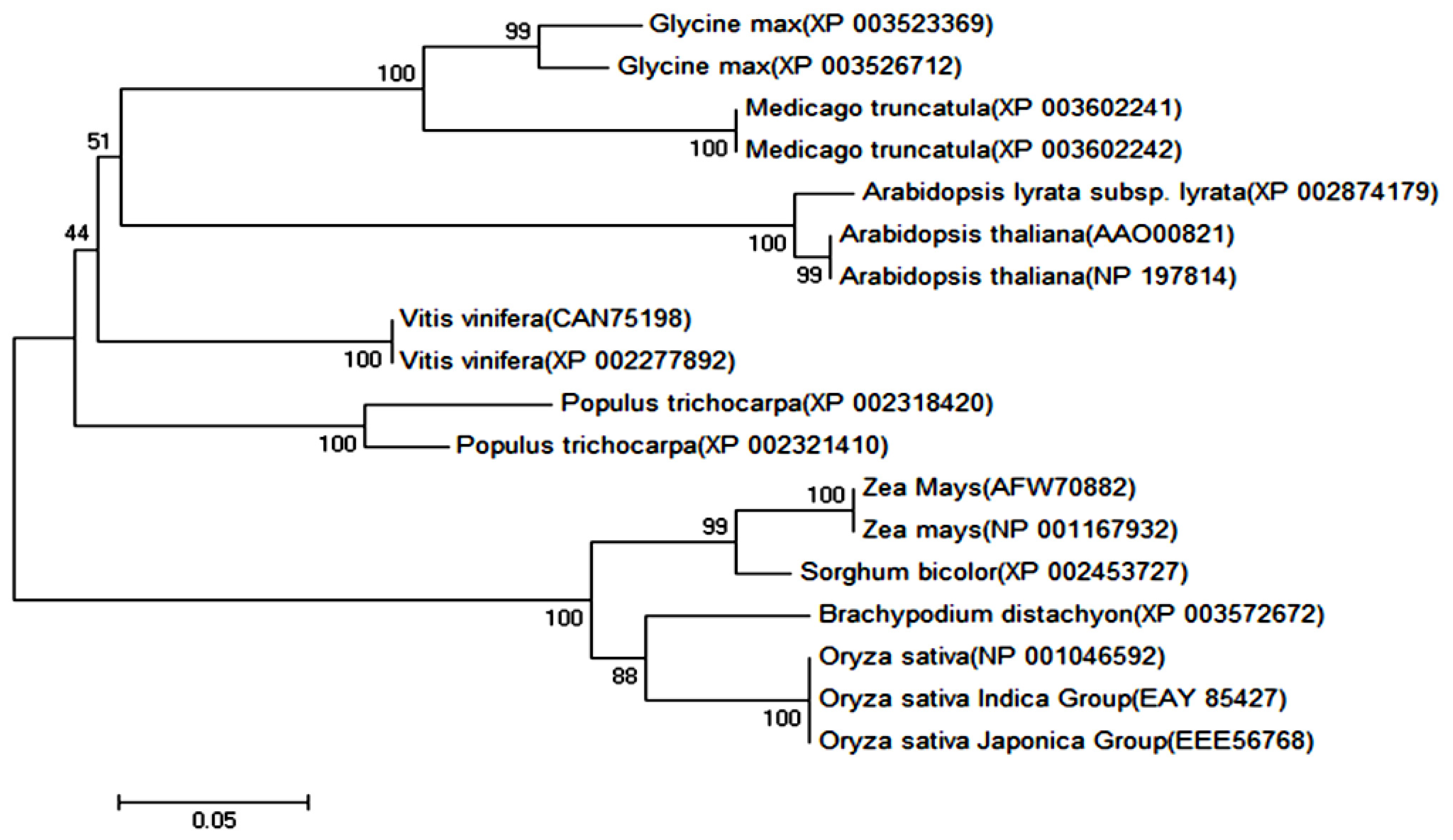
2.1.1. Cloning and Sequence Analysis of OsPOP5 Gene
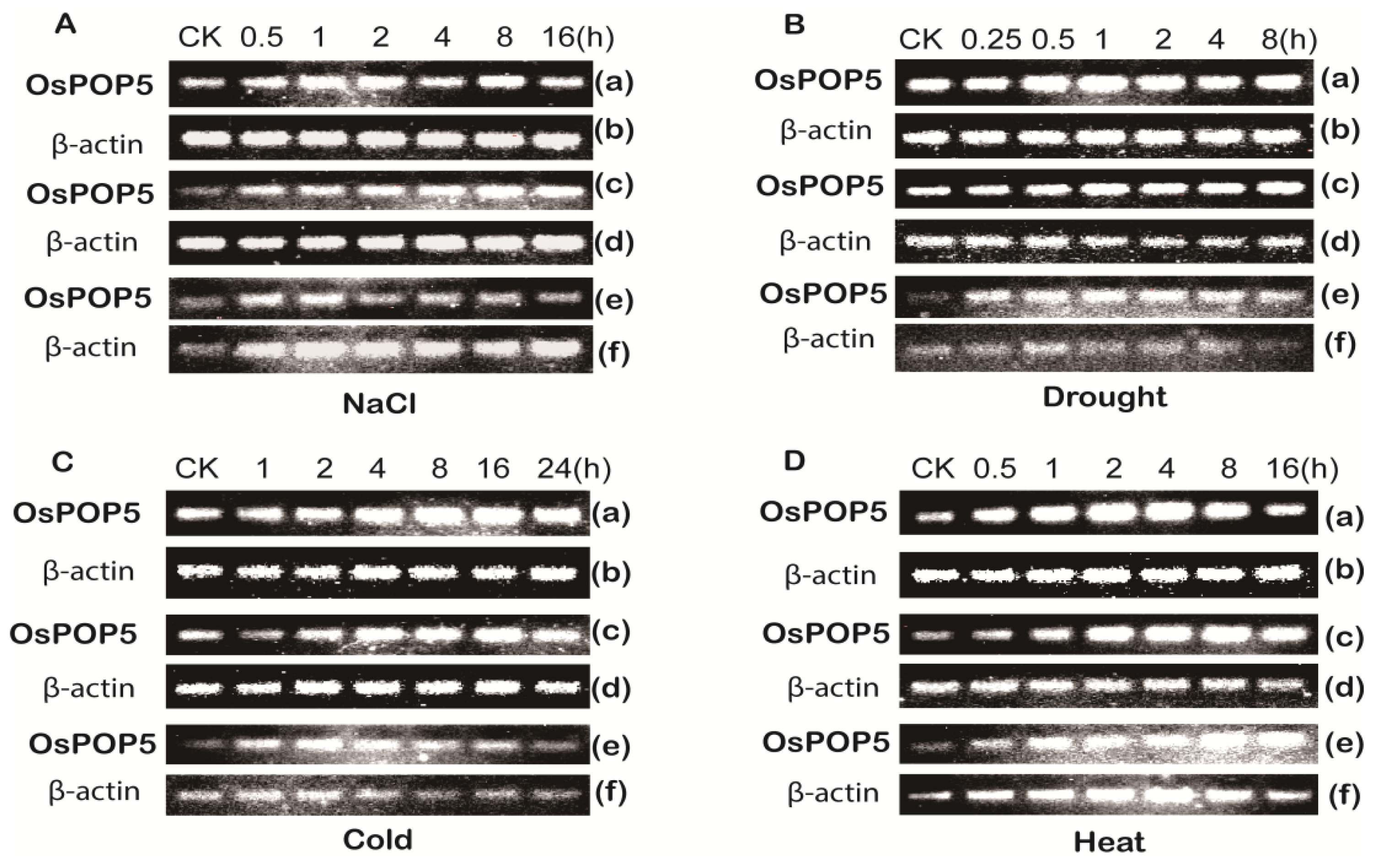
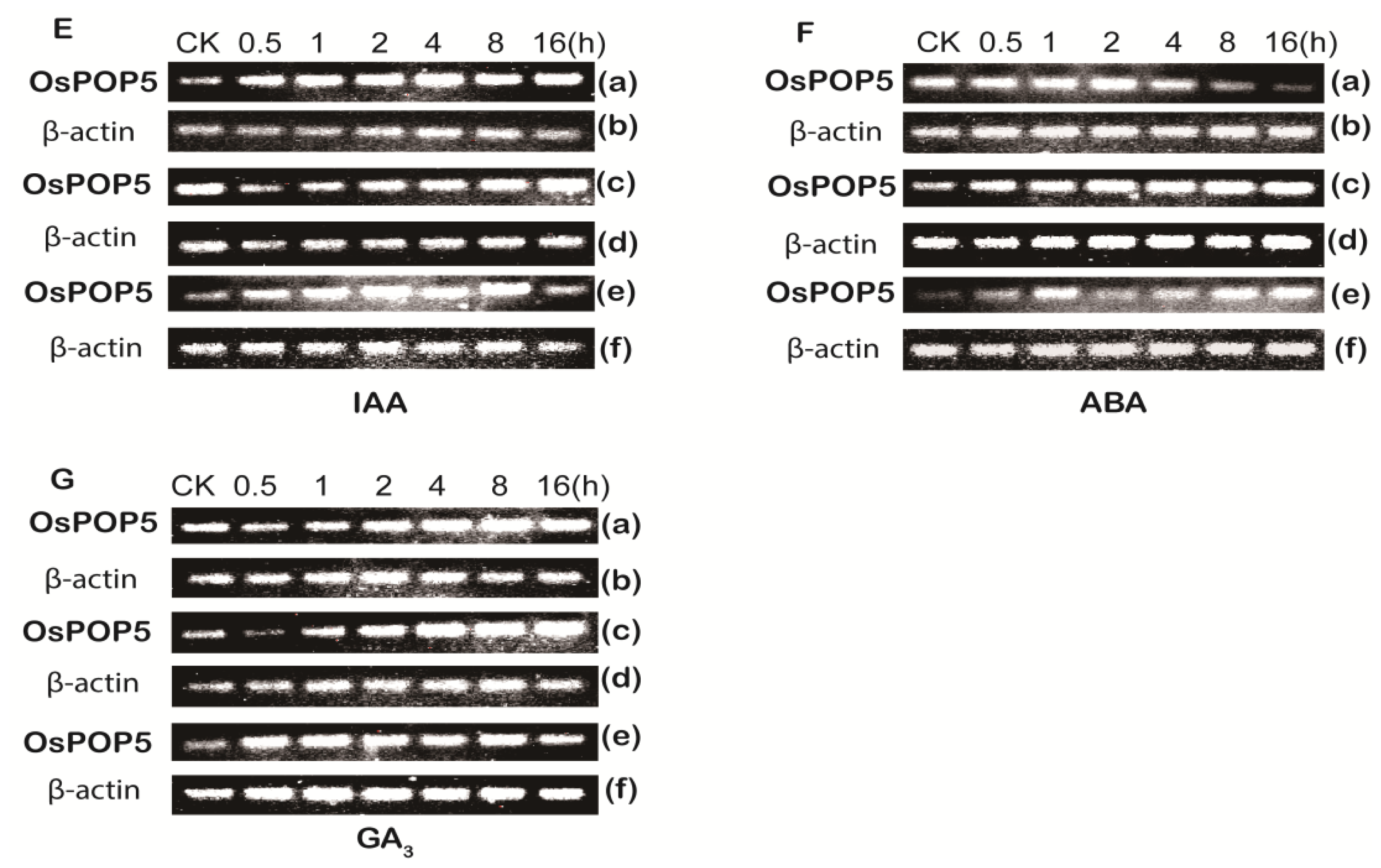
2.1.2. Expression of OsPOP5 under Different Stress Treatments
2.1.3. Expression Analysis of OsPOP5 in E. coli by SDS-PAGE
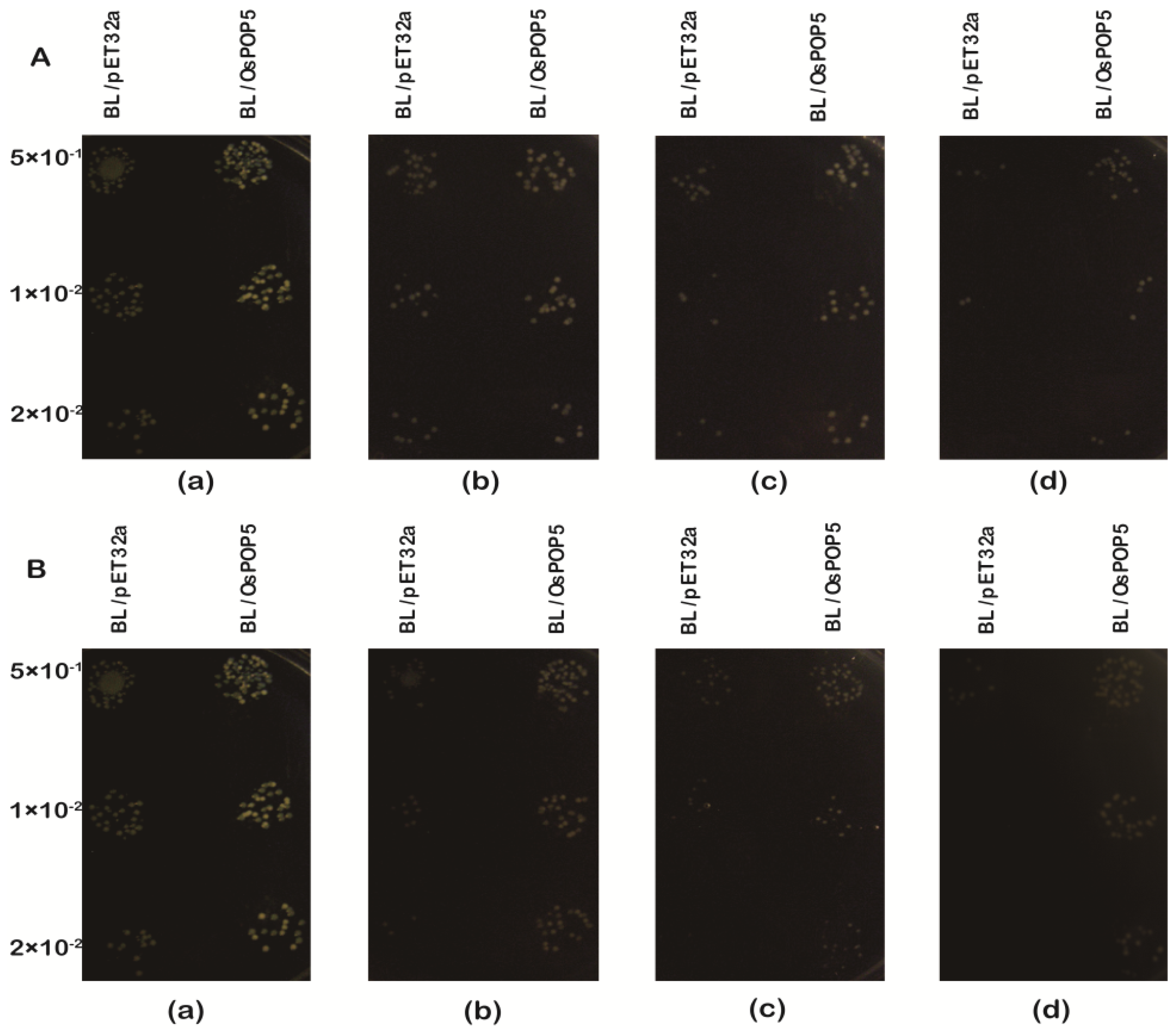
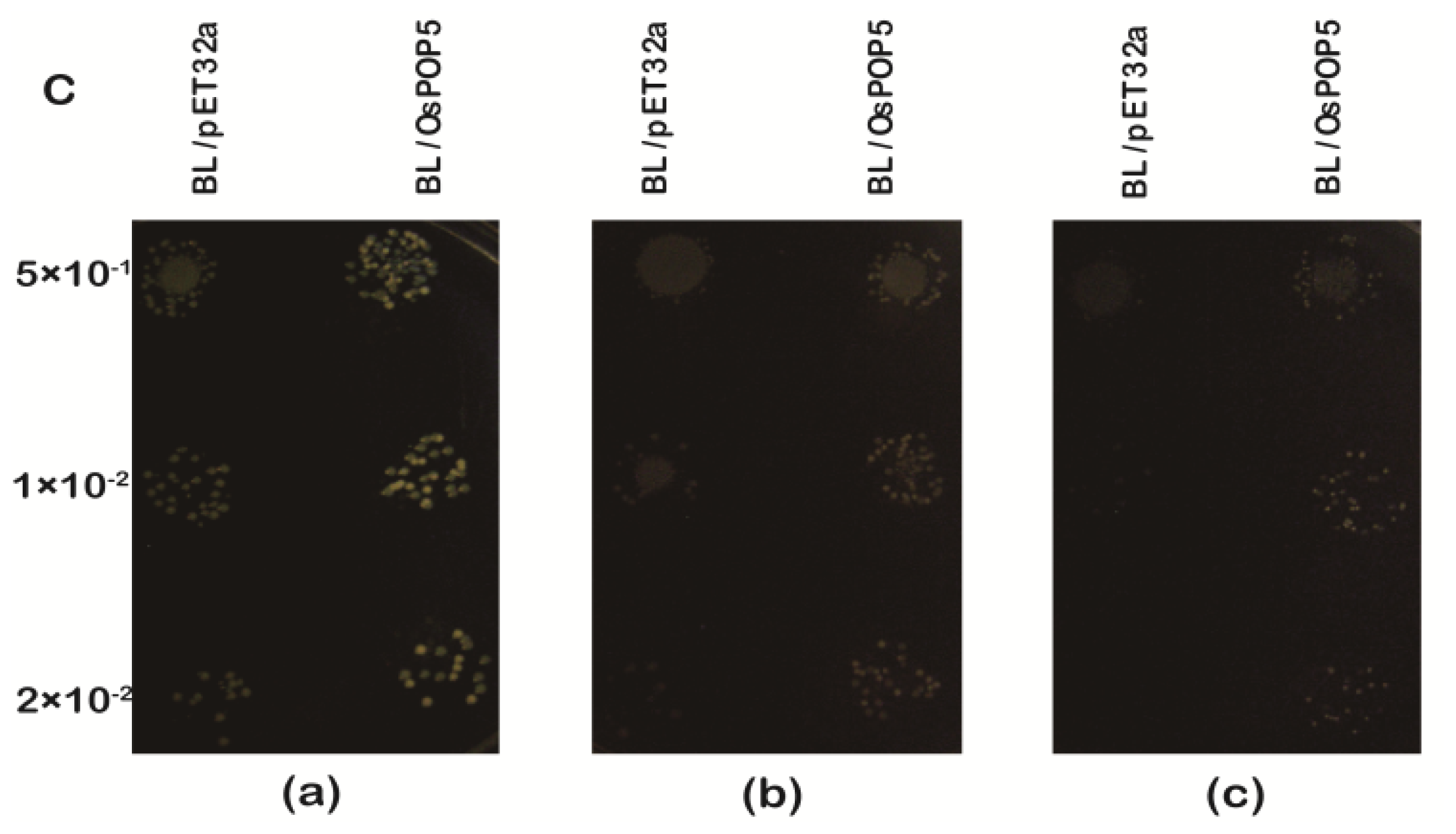
2.1.4. Overexpression of OsPOP5 in E. coli Enhances Growth during Abiotic Stresses
2.2. Discussion
3. Experimental Section
3.1. Plant Material and Abiotic Treatments
3.2. RNA Isolation and Reverse Transcription (RT)
3.3. Isolation and Sequence Analysis of OsPOP5
3.4. Semi-Quantitative PCR Analyse
3.5. Construction of OsPOP5 Expression Vector
3.6. OsPOP5 Expression in E. coli BL21
3.7. Assays for Abiotic Stress Tolerance of E. coli Transformants
4. Conclusions


Acknowledgments
Conflicts of Interest
References
- Yanez, M.; Caceres, S.; Orellana, S.; Bastias, A.; Verdugo, I.; Ruiz-Lara, S.; Casaretto, J.A. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep 2009, 28, 1497–1507. [Google Scholar]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar]
- Gilroy, E.M.; Hein, I.; van der Hoorn, R.; Boevink, P.C.; Venter, E.; McLellan, H.; Kaffarnik, F.; Hrubikova, K.; Shaw, J.; Holeva, M.; et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. Cell Mol. Biol 2007, 52, 1–13. [Google Scholar]
- Parrott, D.; Yang, L.; Shama, L.; Fischer, A.M. Senescence is accelerated, and several proteases are induced by carbon “feast” conditions in barley (Hordeum vulgare L.) leaves. Planta 2005, 222, 989–1000. [Google Scholar]
- Polgar, L. Prolyl oligopeptidases. Methods Enzymol 1994, 244, 188–200. [Google Scholar]
- Polgar, L. The prolyl oligopeptidase family. Cell. Mol. Life Sci. CMLS 2002, 59, 349–362. [Google Scholar]
- Rawlings, N.; Polgar, L.; Barrett, A. A new family of serine-type peptidases related to prolyl oligopeptidase. Biochem. J 1991, 279, 907–908. [Google Scholar]
- Tsuji, A.; Fujisawa, Y.; Mino, T.; Yuasa, K. Identification of a plant aminopeptidase with preference for aromatic amino acid residues as a novel member of the prolyl oligopeptidase family of serine proteases. J. Biochem 2011, 150, 525–534. [Google Scholar]
- Rea, D.; Fülöp, V. Structure-function properties of prolyl oligopeptidase family enzymes. Cell Biochem. Biophys 2006, 44, 349–365. [Google Scholar]
- Venäläinen, J.I.; Juvonen, R.O.; Männistö, P.T. Evolutionary relationships of the prolyl oligopeptidase family enzymes. Eur. J. Biochem 2004, 271, 2705–2715. [Google Scholar]
- Barrett, A.J.; Rawlings, N.D. Families and clans of serine peptidases. Arch. Biochem. Biophys 1995, 318, 247–250. [Google Scholar]
- Rawlings, N.D.; Barrett, A.J. Families of serine peptidases. Methods Enzymol 1994, 244, 19–61. [Google Scholar]
- Polgár, L. Structural relationship between lipases and peptidases of the prolyl oligopeptidase family. FEBS Lett 1992, 311, 281–284. [Google Scholar]
- Goossens, F.; Meester, I.; Vanhoof, G.; Hendriks, D.; Vriend, G.; Scharpé, S. The purification, characterization and analysis of primary and secondary-structure of prolyl oligopeptidase from human lymphocytes. Eur. J. Biochem 1995, 233, 432–441. [Google Scholar]
- Fülöp, V.; Böcskei, Z.; Polgár, L. Prolyl oligopeptidase: An unusual β-propeller domain regulates proteolysis. Cell 1998, 94, 161–170. [Google Scholar]
- Koida, M.; Walter, R. Post-proline cleaving enzyme. Purification of this endopeptidase by affinity chromatography. J. Biol. Chem 1976, 251, 7593–7599. [Google Scholar]
- Walter, R.; Shlank, H.; Glass, J.; Schwartz, I.; Kerenyi, T. Leucylglycinamide released from oxytocin by human uterine enzyme. Science 1971, 173, 827–829. [Google Scholar]
- Rennex, D.; Hemmings, B.A.; Hofsteenge, J.; Stone, S.R. cDNA cloning of porcine brain prolyl endopeptidase and identification of the active-site seryl residue. Biochemistry 1991, 30, 2195–2203. [Google Scholar]
- Vanhoof, G.; Goossens, F.; Hendriks, L.; de Meester, I.; Hendriks, D.; Vriend, G.; van Broeckhoven, C.; Scharpé, S. Cloning and sequence analysis of the gene encoding human lymphocyte prolyl endopeptidase. Gene 1994, 149, 363–366. [Google Scholar]
- Ishino, T.; Ohtsuki, S.; Homma, K.; Natori, S. cDNA cloning of mouse prolyl endopeptidase and its involvement in DNA synthesis by Swiss 3T3 cells. J. Biochem 1998, 123, 540–545. [Google Scholar]
- Yoshimoto, T.; Miyazaki, K.; Haraguchi, N.; Kitazono, A.; Kabashima, T.; Ito, K. Cloning and expression of the cDNA encoding prolyl oligopeptidase (prolyl endopeptidase) from bovine brain. Biol. Pharm. Bull 1997, 20, 1047–1050. [Google Scholar]
- Kimura, A.; Yoshida, I.; Takagi, N.; Takahashi, T. Structure and localization of the mouse prolyl oligopeptidase gene. J. Biol. Chem 1999, 274, 24047–24053. [Google Scholar]
- Pacaud, M.; Richaud, C. Protease II from Escherichia coli. Purification and characterization. J. Biol. Chem 1975, 250, 7771–7779. [Google Scholar]
- Caler, E.V.; Morty, R.E.; Burleigh, B.A.; Andrews, N.W. Dual role of signaling pathways leading to Ca2+ and cyclic AMP elevation in host cell invasion by Trypanosoma cruzi. Infect. Immun 2000, 68, 6602–6610. [Google Scholar]
- Caler, E.V.; de Avalos, S.V.; Haynes, P.A.; Andrews, N.W.; Burleigh, B.A. Oligopeptidase B-dependent signaling mediates host cell invasion by Trypanosoma cruzi. EMBO J 1998, 17, 4975–4986. [Google Scholar]
- Cunningham, D.F.; O’Connor, B. Proline specific peptidases. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol 1997, 1343, 160–186. [Google Scholar]
- Hartel, S.; Gossrau, R.; Hanski, C.; Reutter, W. Dipeptidyl peptidase (DPP) IV in rat organs. Histochemistry 1988, 89, 151–161. [Google Scholar]
- NCBI (National Center for Biotechnology Information). Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 20 June 2013).
- ClustalW. Available online: http://www.ebi.ac.uk/clustalw/ (accessed on 25 June 2013).
- Abogadallah, G.M.; Nada, R.M.; Malinowski, R.; Quick, P. Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta 2011, 233, 1265–1276. [Google Scholar]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 2008, 49, 1135–1149. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 2005, 10, 88–94. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol 2006, 57, 781–803. [Google Scholar]
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 2003, 132, 1020–1032. [Google Scholar]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci 2000, 5, 199–206. [Google Scholar]
- Dracup, M.; Turner, N.; Tang, C.; Reader, M.; Palta, J.; Gladstones, J.; Atkins, C.; Hamblin, J. Responses to abiotic stresses. Lupins Crop Plants Biol. Prod. Util 1998, 227–262. [Google Scholar]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol 2003, 6, 410–417. [Google Scholar]
- Mentlein, R. Proline residues in the maturation and degradation of peptide hormones and neuropeptides. FEBS Lett 1988, 234, 251–256. [Google Scholar]
- Wilk, S. Prolyl endopeptidase. Life Sci 1983, 33, 2149–2157. [Google Scholar]
- Williams, R.; Eames, M.; Ryves, W.; Viggars, J.; Harwood, A. Loss of a prolyl oligopeptidase confers resistance to lithium by elevation of inositol (1,4,5) trisphosphate. EMBO J 1999, 18, 2734–2745. [Google Scholar]
- Schulz, I.; Zeitschel, U.; Rudolph, T.; Ruiz-Carrillo, D.; Rahfeld, J.U.; Gerhartz, B.; Bigl, V.; Demuth, H.U.; Roßner, S. Subcellular localization suggests novel functions for prolyl endopeptidase in protein secretion. J. Neurochem 2005, 94, 970–979. [Google Scholar]
- Sakaguchi, M.; Matsuda, T.; Matsumura, E.; Yoshimoto, T.; Takaoka, M. Prolyl oligopeptidase participates in cell cycle progression in a human neuroblastoma cell line. Biochem. Biophys. Res. Commun 2011, 409, 693–698. [Google Scholar]
- Myöhänen, T.T.; Tenorio-Laranga, J.; Jokinen, B.; Vázquez-Sánchez, R.; Moreno-Baylach, M.; García-Horsman, J.; Männistö, P. Prolyl oligopeptidase induces angiogenesis both in vitro and in vivo in a novel regulatory manner. Br. J. Pharmacol 2011, 163, 1666–1678. [Google Scholar]
- Moreno-Baylach, M.J.; Puttonen, K.A.; Tenorio-Laranga, J.; Venäläinen, J.I.; Storvik, M.; Forsberg, M.M.; García-Horsman, J.A. Prolyl endopeptidase is involved in cellular signalling in human neuroblastoma SH-SY5Y cells. Neurosignals 2011, 19, 97–109. [Google Scholar]
- Agirregoitia, N.; Irazusta, A.; Ruiz, F.; Irazusta, J.; Gil, J. Ontogeny of soluble and particulate prolyl endopeptidase activity in several areas of the rat brain and in the pituitary gland. Dev. Neurosci 2003, 25, 316–323. [Google Scholar]
- Myöhänen, T.T.; Venäläinen, J.I.; Garcia-Horsman, J.A.; Piltonen, M.; Männistö, P.T. Cellular and subcellular distribution of rat brain prolyl oligopeptidase and its association with specific neuronal neurotransmitters. J. Comp. Neurol 2008, 507, 1694–1708. [Google Scholar]
- Myöhänen, T.T.; Venäläinen, J.I.; Tupala, E.; Garcia-Horsman, J.A.; Miettinen, R.; Männistö, P.T. Distribution of immunoreactive prolyl oligopeptidase in human and rat brain. Neurochem. Res 2007, 32, 1365–1374. [Google Scholar]
- Agirregoitia, N.; Laiz-Carrion, R.; Varona, A.; del Rio, M.M.; Mancera, J.; Irazusta, J. Distribution of peptidase activity in teleost and rat tissues. J. Comp. Physiol. B 2005, 175, 433–444. [Google Scholar]
- Bellemère, G.; Vaudry, H.; Mounien, L.; Boutelet, I.; Jégou, S. Localization of the mRNA encoding prolyl endopeptidase in the rat brain and pituitary. J. Comp. Neurol 2004, 471, 128–143. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tan, C.-M.; Chen, R.-J.; Zhang, J.-H.; Gao, X.-L.; Li, L.-H.; Wang, P.-R.; Deng, X.-J.; Xu, Z.-J. OsPOP5, A Prolyl Oligopeptidase Family Gene from Rice Confers Abiotic Stress Tolerance in Escherichia coli. Int. J. Mol. Sci. 2013, 14, 20204-20219. https://doi.org/10.3390/ijms141020204
Tan C-M, Chen R-J, Zhang J-H, Gao X-L, Li L-H, Wang P-R, Deng X-J, Xu Z-J. OsPOP5, A Prolyl Oligopeptidase Family Gene from Rice Confers Abiotic Stress Tolerance in Escherichia coli. International Journal of Molecular Sciences. 2013; 14(10):20204-20219. https://doi.org/10.3390/ijms141020204
Chicago/Turabian StyleTan, Cun-Mei, Rong-Jun Chen, Jian-Hua Zhang, Xiao-Ling Gao, Li-Hua Li, Ping-Rong Wang, Xiao-Jian Deng, and Zheng-Jun Xu. 2013. "OsPOP5, A Prolyl Oligopeptidase Family Gene from Rice Confers Abiotic Stress Tolerance in Escherichia coli" International Journal of Molecular Sciences 14, no. 10: 20204-20219. https://doi.org/10.3390/ijms141020204



