The Role of Vascular Endothelial Growth Factor A Polymorphisms in Breast Cancer
Abstract
:1. Breast Cancer
2. Angiogenesis in Breast Cancer
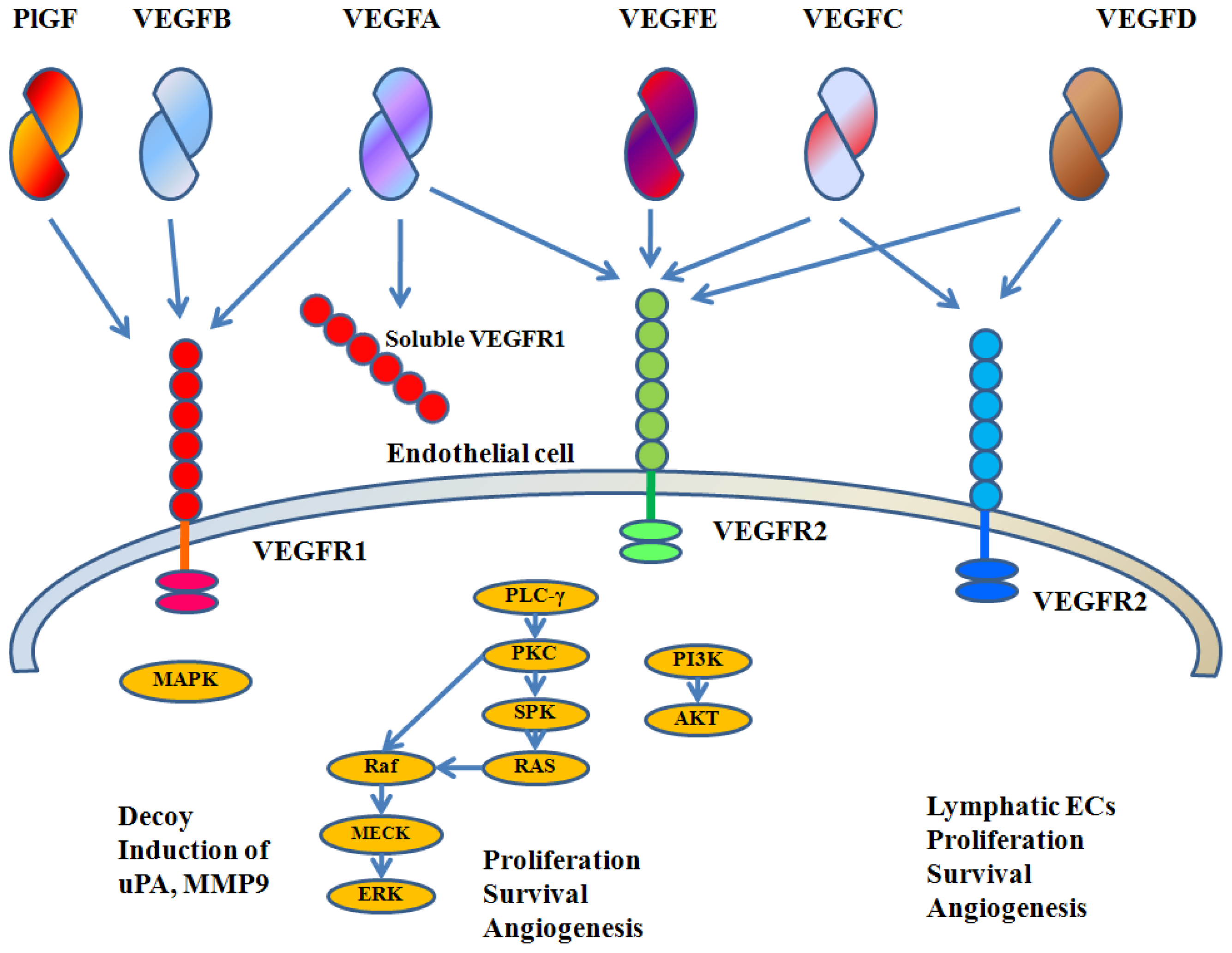
3. VEGFA Biology
4. Roles of VEGFA in Breast Cancer
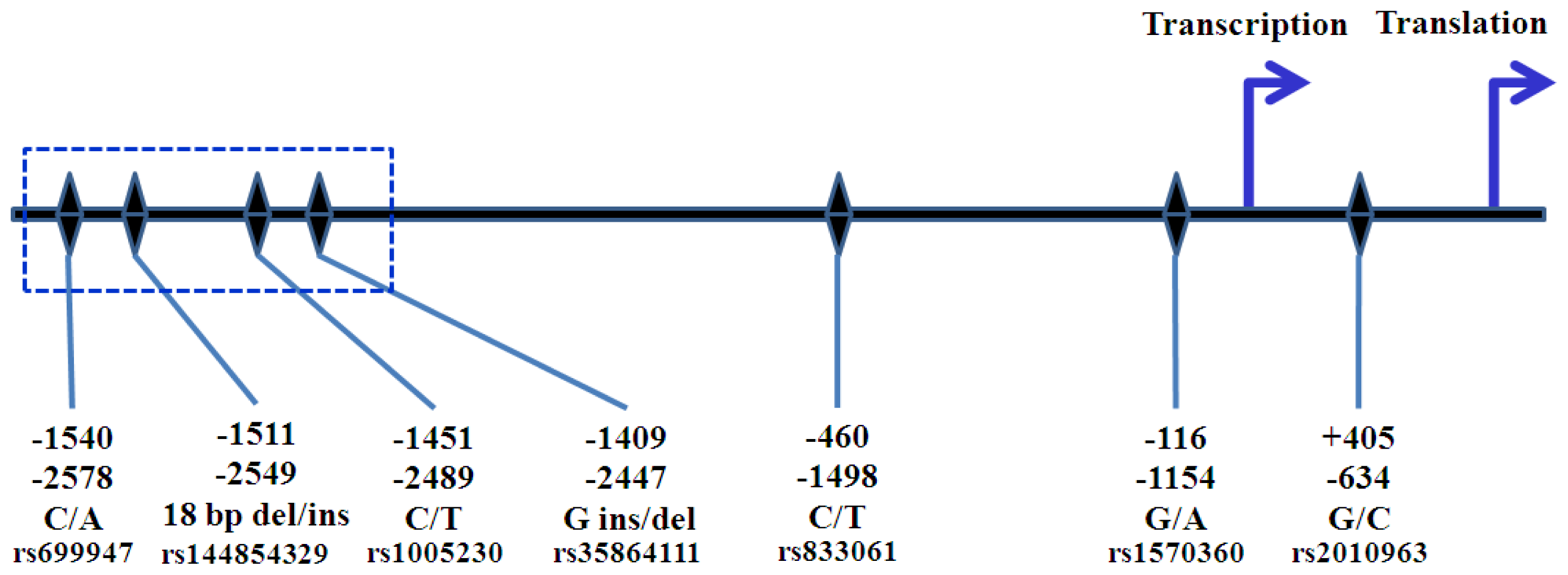
5. VEGFA Polymorphisms
6. The Role of VEGFA Polymorphisms in Breast Cancer Risk and Aggressiveness
7. Potential Clinical Application of VEGFA Polymorphisms
Acknowledgments
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin 2011, 61, 69–90. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Bergers, G.; Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev 2003, 3, 401–410. [Google Scholar]
- Jain, R.K. Determinants of tumor blood flow: A review. Cancer Res 1988, 48, 2641–2658. [Google Scholar]
- Jain, R.K.; Munn, L.L.; Fukumura, D. Dissecting tumour pathophysiology using intravital microscopy. Nat. Rev 2002, 2, 266–276. [Google Scholar]
- Anderson, S.A.; Glod, J.; Arbab, A.S.; Noel, M.; Ashari, P.; Fine, H.A.; Frank, J.A. Noninvasive MR imaging of magnetically labeled stem cells to directly identify neovasculature in a glioma model. Blood 2005, 105, 420–425. [Google Scholar]
- Djonov, V.; Andres, A.-C.; Ziemiecki, A. Vascular remodelling during the normal and malignant life cycle of the mammary gland. Microsc. Res. Tech 2001, 52, 182–189. [Google Scholar]
- Brem, S.S.; Gullino, P.M.; Medina, D. Angiogenesis: A marker for neoplastic transformation of mammary papillary hyperplasia. Science 1977, 195, 880–882. [Google Scholar]
- Jensen, H.M.; Chen, I.; DeVault, M.R.; Lewis, A.E. Angiogenesis induced by “normal” human breast tissue: a probable marker for precancer. Science 1982, 218, 293–295. [Google Scholar]
- Guinebretiere, J.M.; le Monique, G.; Gavoille, A.; Bahi, J.; Contesso, G. Angiogenesis and risk of breast cancer in women with fibrocystic disease. J. Natl. Cancer Inst 1994, 86, 635–636. [Google Scholar]
- Weidner, N.; Folkman, J.; Pozza, F.; Bevilacqua, P.; Allred, E.N.; Moore, D.H.; Meli, S.; Gasparini, G. Tumor angiogenesis: A new significant and independent prognostic indicator in early-stage breast carcinoma. J. Natl. Cancer Inst 1992, 84, 1875–1887. [Google Scholar]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor angiogenesis and metastasis—correlation in invasive breast carcinoma. N. Engl. J. Med 1991, 324, 1–8. [Google Scholar]
- Uzzan, B.; Nicolas, P.; Cucherat, M.; Perret, G.Y. Microvessel density as a prognostic factor in women with breast cancer: a systematic review of the literature and meta-analysis. Cancer Res 2004, 64, 2941–2955. [Google Scholar]
- Vincenti, V.; Cassano, C.; Rocchi, M.; Persico, G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation 1996, 93, 1493–1495. [Google Scholar]
- Houck, K.A. The vascular endothelial growth factor family: Identification of a fourth molecular species and characterization of alternative splicing of RNA. Mol. Endocrinol 1991, 5, 1806–1814. [Google Scholar]
- Tischer, E. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem 1991, 266, 11947–11954. [Google Scholar]
- Neufeld, G.; Cohen, T.; Gengrinovitch, S.; Poltorak, Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J 1999, 13, 9–22. [Google Scholar]
- Leung, D.W.; Cachianes, G.; Kuang, W.J.; Goeddel, D.V.; Ferrara, N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 1989, 246, 1306–1309. [Google Scholar]
- Plouet, J.; Schilling, J.; Gospodarowicz, D. Isolation and characterization of a newly identified endothelial cell mitogen produced by AtT20 cells. EMBO J 1989, 8, 3801–3808. [Google Scholar]
- Nagy, J.A. Vascular permeability factor/vascular endothelial growth factor induces lymphangiogenesis as well as angiogenesis. J. Exp. Med 2002, 196, 1497–1506. [Google Scholar]
- Matsumoto, T.; Claesson-Welsh, L. VEGF receptor signal transduction. Science STKE 2001, 112, 1–17. [Google Scholar]
- Gerber, H.P. VEGF regulates endothelial cell survival by the PI3-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J. Biol. Chem 1998, 273, 30366–30343. [Google Scholar]
- Gerber, H.P.; Dixit, V.; Ferrara, N. Vascular endothelial growth factor induces expression of the antiapoptotic proteins Bcl-2 and A1 in vascular endothelial cells. J. Biol. Chem 1998, 273, 13313–13316. [Google Scholar]
- Gerber, H.P. VEGF is required for growth and survival in neonatal mice. Development 1999, 126, 1149–1159. [Google Scholar]
- Benjamin, L.E.; Golijanin, D.; Itin, A.; Pode, D.; Keshet, E. Selective ablation of immature blood vessels in established human tumors follows vascular endothelial growth factor withdrawal. J. Clin. Invest 1999, 103, 159–165. [Google Scholar]
- Yuan, F. Time-dependent vascular regression and permeability changes in established human tumor xenografts induced by an anti-vascular endothelial growth factor/vascular permeability factor antibody. Proc. Natl. Acad. Sci. USA 1996, 93, 14765–14770. [Google Scholar]
- Senger, D.R. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science 1983, 219, 983–985. [Google Scholar]
- Dvorak, H.F.; Brown, L.F.; Detmar, M.; Dvorak, A.M. Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am. J. Pathol 1995, 146, 1029–1039. [Google Scholar]
- Bates, D.O.; Curry, F.E. Vascular endothelial growth factor increases microvascular permeability via a Ca(2+)-dependent pathway. Am. J. Physiol 1997, 273, H687–H694. [Google Scholar]
- Roberts, W.G.; Palade, G.E. Increased microvascular permeability and endothelial fenestration induced by vascular endothelial growth factor. J. Cell. Sci 1995, 108, 2369–2379. [Google Scholar]
- Ku, D.D.; Zaleski, J.K.; Liu, S.; Brock, T.A. Vascular endothelial growth factor induces EDRF-dependent relaxation in coronary arteries. Am. J. Physiol 1993, 265, H586–H592. [Google Scholar]
- Yang, R. Effects of vascular endothelial growth factor on hemodynamics and cardiac performance. J. Cardiovasc. Pharmacol 1996, 27, 838–844. [Google Scholar]
- Kim, K.J. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumor growth in vivo. Nature 1993, 362, 841–844. [Google Scholar]
- Ferrara, N.; Davis-Smyth, T. The biology of vascular endothelial growth factor. Endocr. Rev 1997, 18, 4–25. [Google Scholar]
- Fukumura, D. Tumor induction of VEGF promoter activity in stromal cells. Cell 1998, 94, 715–725. [Google Scholar]
- Gerber, H.P.; Kowalski, J.; Sherman, D.; Eberhard, D.A.; Ferrara, N. Complete inhibition of rhabdomyosarcoma xenograft growth and neovascularization requires blockade of both tumor and host vascular endothelial growth factor. Cancer Res 2000, 60, 6253–6258. [Google Scholar]
- Tsuzuki, Y. Vascular endothelial growth factor (VEGF) modulation by targeting hypoxia-inducible factor-1[alpha→hypoxia response element→VEGF cascade differ entially regulates vascular response and growth rate in tumors. Cancer Res 2000, 60, 6248–6252. [Google Scholar]
- Bergers, G. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell. Biol 2000, 2, 737–744. [Google Scholar]
- Gasparini, G. Prognostic value of vascular endothelial growth factor in breast cancer. Oncologist 2000, 5, 37–44. [Google Scholar]
- Salven, P.; Perhoniemi, V.; Tykka, H.; Maenpaa, H.; Joensuu, H. Serum VEGF levels in women with a benign breast tumor or breast cancer. Breast Cancer Res. Treat 1999, 53, 161–166. [Google Scholar]
- Obermair, A.; Kucera, E.; Mayerhofer, K.; Speiser, P.; Seifert, M.; Czerwenka, K.; Kaider, A.; Leodolter, S.; Kainz, C.; Zeillinger, R. Vascular endothelial growth factor (VEGF) in human breast cancer: correlation with disease-free survival. Int. J. Cancer 1997, 74, 455–458. [Google Scholar]
- Ghosh, S.; Sullivan, C.A.; Zerkowski, M.P.; Molinaro, A.M.; Rimm, D.L.; Camp, R.L.; Chung, G.G. High levels of vascular endothelial growth factor and its receptors (VEGFR-1, VEGFR-2, neuropilin-1) are associated with worse outcome in breast cancer. Human Pathol 2008, 39, 1835–1843. [Google Scholar]
- Xie, X.D.; Qu, S.X.; Liu, Z.Z.; Zhang, F.; Zheng, Z.D. Study on relationship between angiogenesis and micrometastases of peripheral blood in breast cancer. J. Cancer Res. Clin 2009, 135, 413–419. [Google Scholar]
- Manders, P.; Beex, L.V.; Tjan-Heijnen, V.C.; Geurts-Moespot, J.; van Tienoven, T.H.; Foekens, J.A.; Sweep, C.G. The prognostic value of vascular endothelial growth factor in 574 node-negative breast cancer patients who did not receive adjuvant systemic therapy. Br. J. Cancer 2002, 87, 772–778. [Google Scholar]
- Bando, H.; Weich, H.A.; Brokelmann, M.; Horiguchi, S.; Funata, N.; Ogawa, T.; Toi, M. Association between intratumoral free and total VEGF, soluble VEGFR-1, VEGFR-2 and prognosis in breast cancer. Br. J. Cancer 2005, 92, 553–561. [Google Scholar]
- Toi, M.; Bando, H.; Ogawa, T.; Muta, M.; Hornig, C.; Weich, H.A. Significance of vascular endothelial growth factor (VEGF)/soluble VEGF receptor-1 relationship in breast cancer. Int. J. Cancer 2002, 98, 14–18. [Google Scholar]
- Ryden, L.; Stendahl, M.; Jonsson, H.; Emdin, S.; Bengtsson, N.O.; Landberg, G. Tumor-specific VEGF-A and VEGFR2 in postmenopausal breast cancer patients with long-term follow-up. Implication of a link between VEGF pathway and tamoxifen response. Breast Cancer Res. Treat 2005, 89, 135–143. [Google Scholar]
- Burstein, H.J.; Chen, Y.H.; Parker, L.M.; Savoie, J.; Younger, J.; Kuter, I.; Ryan, P.D.; Garber, J.E.; Chen, H.; Campos, S.M.; et al. VEGF as a marker for outcome among advanced breast cancer patients receiving anti-VEGF therapy with bevacizumab and vinorelbine chemotherapy. Clin. Cancer Res 2008, 14, 7871–7877. [Google Scholar]
- Brogan, I.J.; Khan, N.; Isaac, K.; Hutchinson, J.A.; Pravica, V.; Hutchinson, I.V. Novel polymorphisms in the promoter and 5′ UTR regions of the human vascular endothelial growth factor gene. Hum. Immunol 1999, 60, 1245–1249. [Google Scholar]
- Watson, C.J.; Webb, N.J.; Bottomley, M.J.; Brenchley, P.E. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: Correlation with variation in VEGF protein production. Cytokine 2000, 12, 1232–1235. [Google Scholar]
- Jin, Q.; Hemminki, K.; Enquist, K.; Lenner, P.; Grzybowska, E.; Klaes, R.; Henriksson, R.; Chen, B.; Pamula, J.; Pekala, W.; et al. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin. Cancer Res 2005, 11, 3647–3653. [Google Scholar]
- Awata, T.; Inoue, K.; Kurihara, S.; Ohkubo, T.; Watanabe, M.; Inukai, K.; Inoue, I.; Katayama, S. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes 2002, 51, 1635–1639. [Google Scholar]
- Shahbazi, M.; Fryer, A.A.; Pravica, V.; Brogan, I.J.; Ramsay, H.M.; Hutchinson, I.V.; Harden, P.N. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J. Am. Soc. Nephrol 2002, 13, 260–264. [Google Scholar]
- Koukourakis, M.I.; Papazoglou, D.; Giatromanolaki, A.; Bougioukas, G.; Maltezos, E.; Sivridis, E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer 2004, 46, 293–298. [Google Scholar]
- Stevens, A.; Soden, J.; Brenchley, P.E.; Ralph, S.; Ray, D.W. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res 2003, 63, 812–816. [Google Scholar]
- Pages, G.; Pouyssegur, J. Transcriptional regulation of the Vascular Endothelial Growth Factor gene—a concert of activating factors. Cardiovasc. Res 2005, 65, 564–573. [Google Scholar]
- Morris, J.F.; Hromas, R.; Rauscher, F.J., III. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol. Cell. Biol. 1994, 14, 1786–1795. [Google Scholar]
- Awata, T.; Kurihara, S.; Takata, N.; Neda, T.; Iizuka, H.; Ohkubo, T.; Osaki, M.; Watanabe, M.; Nakashima, Y.; Inukai, K.; et al. Functional VEGF C-634G polymorphism is associated with development of diabetic macular edema and correlated with macular retinal thickness in type 2 diabetes. Biochem. Biophys. Res. Commun 2005, 333, 679–685. [Google Scholar]
- Huez, I.; Bornes, S.; Bresson, D.; Creancier, L.; Prats, H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol. Endocrinol 2001, 15, 2197–2210. [Google Scholar]
- Krippl, P.; Langsenlehner, U.; Renner, W.; Yazdani-Biuki, B.; Wolf, G.; Wascher, T.C.; Paulweber, B.; Haas, J.; Samonigg, H. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int. J. Cancer 2003, 106, 468–471. [Google Scholar]
- Renner, W.; Kotschan, S.; Hoffmann, C.; Obermayer-Pietsch, B.; Pilger, E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J. Vasc. Res 2000, 37, 443–448. [Google Scholar]
- Doi, K.; Noiri, E.; Nakao, A.; Fujita, T.; Kobayashi, S.; Tokunaga, K. Functional polymorphisms in the vascular endothelial growth factor gene are associated with development of end-stage renal disease in males. J. Am. Soc. Nephrol 2006, 17, 823–830. [Google Scholar]
- Abe, A.; Sato, K.; Habuchi, T.; Wang, L.; Li, Z.; Tsuchiya, N.; Ohyama, C.; Satoh, S.; Ogawa, O.; Kato, T. Single nucleotide polymorphisms in the 3′ untranslated region of vascular endothelial growth factor gene in Japanese population with or without renal cell carcinoma. Tohoku J. Exp. Med 2002, 198, 181–190. [Google Scholar]
- Garcia-Closas, M.; Malats, N.; Real, F.X.; Yeager, M.; Welch, R.; Silverman, D.; Kogevinas, M.; Dosemeci, M.; Figueroa, J.; Chatterjee, N.; et al. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet 2007, 3, e29. [Google Scholar]
- Jacobs, E.J.; Feigelson, H.S.; Bain, E.B.; Brady, K.A.; Rodriguez, C.; Stevens, V.L.; Patel, A.V.; Thun, M.J.; Calle, E.E. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res 2006, 8, R22. [Google Scholar]
- Schneider, B.P.; Radovich, M.; Sledge, G.W.; Robarge, J.D.; Li, L.; Storniolo, A.M.; Lemler, S.; Nguyen, A.T.; Hancock, B.A.; Stout, M.; et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res. Treat 2008, 111, 157–163. [Google Scholar]
- Kidd, L.R.; Brock, G.N.; VanCleave, T.T.; Benford, M.L.; Lavender, N.A.; Kruer, T.L.; Wittliff, J.L. Angiogenesis-associated sequence variants relative to breast cancer recurrence and survival. Cancer Causes Control 2010, 21, 1545–1557. [Google Scholar]
- Langsenlehner, U.; Wolf, G.; Langsenlehner, T.; Gerger, A.; Hofmann, G.; Clar, H.; Wascher, T.C.; Paulweber, B.; Samonigg, H.; Krippl, P.; et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res. Treat 2008, 109, 297–304. [Google Scholar]
- Balasubramanian, S.P.; Cox, A.; Cross, S.S.; Higham, S.E.; Brown, N.J.; Reed, M.W. Influence of VEGF-A gene variation and protein levels in breast cancer susceptibility and severity. Int. J. Cancer 2007, 121, 1009–1016. [Google Scholar]
- Kataoka, N.; Cai, Q.; Wen, W.; Shu, X.O.; Jin, F.; Gao, Y.T.; Zheng, W. Population-based case-control study of VEGF gene polymorphisms and breast cancer risk among Chinese women. Cancer Epidemiol. Biomark. Prev 2006, 15, 1148–1152. [Google Scholar]
- Smith, K.C.; Bateman, A.C.; Fussell, H.M.; Howell, W.M. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur. J. Immunogenet 2004, 31, 167–173. [Google Scholar]
- Oliveira, C.; Lourenco, G.J.; Silva, P.M.; Cardoso-Filho, C.; Favarelli, M.H.; Goncales, N.S.; Gurgel, M.S.; Lima, C.S. Polymorphisms in the 5′- and 3′-untranslated region of the VEGF gene and sporadic breast cancer risk and clinicopathologic characteristics. Tumour Biol 2011, 32, 295–300. [Google Scholar]
- Beeghly-Fadiel, A.; Shu, X.O.; Lu, W.; Long, J.; Cai, Q.; Xiang, Y.B.; Zheng, Y.; Zhao, Z.; Gu, K.; Gao, Y.T.; et al. Genetic variation in VEGF family genes and breast cancer risk: A report from the Shanghai Breast Cancer Genetics Study. Cancer Epidemiol. Biomark. Prev 2011, 20, 33–41. [Google Scholar]
- Eroglu, A.; Ozturk, A.; Cam, R.; Akar, N. Vascular endothelial growth factor gene 936 C/T polymorphism in breast cancer patients. Med. Oncol 2008, 25, 54–55. [Google Scholar]
- Gerger, A.; Langsenlehner, U.; Renner, W.; Weitzer, W.; Eder, T.; Yazdani-Biuki, B.; Hofmann, G.; Samonigg, H.; Krippl, P. A multigenic approach to predict breast cancer risk. Breast Cancer Res. Treat 2007, 104, 159–164. [Google Scholar]
- Jakubowska, A.; Gronwald, J.; Menkiszak, J.; Gorski, B.; Huzarski, T.; Byrski, T.; Edler, L.; Lubinski, J.; Scott, R.J.; Hamann, U. The VEGF_936_C>T 3′UTR polymorphism reduces BRCA1-associated breast cancer risk in Polish women. Cancer Lett 2008, 262, 71–76. [Google Scholar]
- Jakubowska, A.; Jaworska, K.; Cybulski, C.; Janicka, A.; Szymanska-Pasternak, J.; Lener, M.; Narod, S.A.; Lubinski, J. Do BRCA1 modifiers also affect the risk of breast cancer in non-carriers? Eur. J. Cancer 2009, 45, 837–842. [Google Scholar]
- Rodrigues, P.; Furriol, J.; Tormo, E.; Ballester, S.; Lluch, A.; Eroles, P. The single-nucleotide polymorphisms +936 C/T VEGF and −710 C/T VEGFR1 are associated with breast cancer protection in a Spanish population. Breast Cancer Res. Treat 2012, 133, 769–778. [Google Scholar]
- Zhang, B.; Beeghly-Fadiel, A.; Lu, W.; Cai, Q.; Xiang, Y.B.; Zheng, Y.; Long, J.; Ye, C.; Gu, K.; Shu, X.O.; et al. Evaluation of functional genetic variants for breast cancer risk: results from the Shanghai breast cancer study. Am. J. Epidemiol 2011, 173, 1159–1170. [Google Scholar]
- Etienne-Grimaldi, M.C.; Formento, P.; Degeorges, A.; Pierga, J.Y.; Delva, R.; Pivot, X.; Dalenc, F.; Espie, M.; Veyret, C.; Formento, J.L.; et al. Prospective analysis of the impact of VEGF-A gene polymorphisms on the pharmacodynamics of bevacizumab-based therapy in metastatic breast cancer patients. Br. J. Clin. Pharmacol 2011, 71, 921–928. [Google Scholar]
- Lu, H.; Shu, X.O.; Cui, Y.; Kataoka, N.; Wen, W.; Cai, Q.; Ruan, Z.X.; Gao, Y.T.; Zheng, W. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res 2005, 65, 5015–5019. [Google Scholar]
- Wolf, G.; Aigner, R.M.; Schaffler, G.; Langsenlehner, U.; Renner, W.; Samonigg, H.; Yazdani-Biuki, B.; Krippl, P. The 936C>T polymorphism of the gene for vascular endothelial growth factor is associated with 18F-fluorodeoxyglucose uptake. Breast Cancer Res. Treat 2004, 88, 205–208. [Google Scholar]
- Knechtel, G.; Hofmann, G.; Gerger, A.; Renner, W.; Langsenlehner, T.; Szkandera, J.; Wolf, G.; Samonigg, H.; Krippl, P.; Langsenlehner, U. Analysis of common germline polymorphisms as prognostic factors in patients with lymph node-positive breast cancer. J. Cancer Res. Clin 2010, 136, 1813–1819. [Google Scholar]
- Wang, K.; Liu, L.; Zhu, Z.M.; Shao, J.H.; Xin, L. Five polymorphisms of vascular endothelial growth factor (VEGF) and risk of breast cancer: a meta-analysis involving 16,703 individuals. Cytokine 2011, 56, 167–173. [Google Scholar]
- Yang, D.S.; Park, K.H.; Woo, O.H.; Woo, S.U.; Kim, A.R.; Lee, E.S.; Lee, J.B.; Kim, Y.H.; Kim, J.S.; Seo, J.H. Association of a vascular endothelial growth factor gene 936 C/T polymorphism with breast cancer risk: a meta-analysis. Breast Cancer Res. Treat 2011, 125, 849–853. [Google Scholar]
- Gu, D.; Wang, M. VEGF 936C>T polymorphism and breast cancer risk: evidence from 5729 cases and 5868 controls. Breast Cancer Res. Treat 2011, 125, 489–493. [Google Scholar]
- Liu, L.; Liu, L.; Zeng, F.; Wang, K.; Huang, J.; Xin, L.; Zhu, P.Q. Meta-analysis of the association between VEGF-634 G>C and risk of malignancy based on 23 case-control studies. J. Cancer Res. Clin 2011, 137, 1027–1036. [Google Scholar]
- Miller, K.; Wang, M.; Gralow, J.; Dickler, M.; Cobleigh, M.; Perez, E.A.; Shenkier, T.; Cella, D.; Davidson, N.E. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N. Engl. J. Med 2007, 357, 2666–2676. [Google Scholar]
- Miles, D.W.; Chan, A.; Dirix, L.Y.; Cortes, J.; Pivot, X.; Tomczak, P.; Delozier, T.; Sohn, J.H.; Provencher, L.; Puglisi, F.; et al. Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J. Clin. Oncol 2010, 28, 3239–3247. [Google Scholar]
- Thomssen, C.; Pierga, J.Y.; Pritchard, K.I.; Biganzoli, L.; Cortes-Funes, H.; Petrakova, K.; Kaufman, B.; Duenne, A.; Smith, I. First-line bevacizumab-containing therapy for triple-negative breast cancer: analysis of 585 patients treated in the ATHENA study. Oncology 2012, 82, 218–227. [Google Scholar]
- Miller, K.D.; Chap, L.I.; Holmes, F.A.; Cobleigh, M.A.; Marcom, P.K.; Fehrenbacher, L.; Dickler, M.; Overmoyer, B.A.; Reimann, J.D.; et al. Randomized phase III trial of capecitabine compared with bevacizumab plus capecitabine in patients with previously treated metastatic breast cancer. J. Clin. Oncol 2005, 23, 792–799. [Google Scholar]
- Kostopoulos, I.; Arapantoni-Dadioti, P.; Gogas, H.; Papadopoulos, S.; Malamou-Mitsi, V.; Scopa, C.D.; Markaki, S.; Karagianni, E.; Kyriakou, V.; Margariti, A.; et al. Evaluation of the prognostic value of HER-2 and VEGF in breast cancer patients participating in a randomized study with dose-dense sequential adjuvant chemotherapy. Breast Cancer Res. Treat 2006, 96, 251–261. [Google Scholar]
- Ludovini, V.; Sidoni, A.; Pistola, L.; Bellezza, G.; de Angelis, V.; Gori, S.; Mosconi, A.M.; Bisagni, G.; Cherubini, R.; Bian, A.R.; et al. Evaluation of the prognostic role of vascular endothelial growth factor and microvessel density in stages I and II breast cancer patients. Breast Cancer Res. Treat 2003, 81, 159–168. [Google Scholar]
- MacConmara, M.; O’Hanlon, D.M.; Kiely, M.J.; Connolly, Y.; Jeffers, M.; Keane, F.B. An evaluation of the prognostic significance of vascular endothelial growth factor in node positive primary breast carcinoma. Int. J. Oncol 2002, 20, 717–721. [Google Scholar]
- Schneider, B.P.; Wang, M.; Radovich, M.; Sledge, G.W.; Badve, S.; Thor, A.; Flockhart, D.A.; Hancock, B.; Davidson, N.; Gralow, J.; et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J. Clin. Oncol 2008, 26, 4672–4678. [Google Scholar]
| SNPs | Authors | Population | Case/control | Susceptibility |
|---|---|---|---|---|
| −2578C/A rs699947 | Langsenlehner et al.[68] | Austrian | 804/804 | N.S. |
| Jacobs et al.[65] | American | 504/501 | C allele was associated with increased risk of invasive cancer, OR = 1.46 (1.00–2.14), p (trend) = 0.049. | |
| Jin et al.[51] | Polish German Swedish | 1525/1503 | N.S. | |
| Schneider et al.[66] | Caucasian, African- American | 175/520 | AA genotype was associated with higher risk of breast cancer, OR = 1.99 (1.06–3.74), p = 0.03 (adjusted by Gail score). | |
| −2489C/T rs1005230 | Langsenlehner et al.[68] | Austrian | 804/804 | N.S. |
| −1498C/T rs833061 | Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Kataoka et al.[70] | Chinese | 1184/1093 | N.S. | |
| Schneider et al.[66] | Caucasian, African- American | 175/520 | CC genotype was associated with higher risk of breast cancer, OR = 2.01 (1.08–3.76), p = 0.03. | |
| −1154G/A rs1570360 | Jacobs et al.[65] | American | 504/501 | G allele was associated with increased risk of invasive cancer, OR = 1.64 (1.02–2.64), p = 0.007. |
| Jin et al.[51] | Polish German | 586/570 | N.S. | |
| Schneider et al.[66] | Caucasian, African-American | 175/520 | N.S. | |
| Smith et al.[71] | English | 263/144 | N.S. | |
| −634G/C rs2010963 | Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Jacobs et al.[65] | American | 504/501 | N.S. | |
| Jin et al.[51] | Swedish | 941/936 | N.S. | |
| Kataoka et al.[70] | Chinese | 1184/1093 | N.S. | |
| Oliveira et al.[72] | Caucasian, African-American | 235/235 | CC genotype was associated with increased risk for breast cancer, OR = 2.20 (1.20–4.02), p = 0.01 when compared to GG and GC genotype. | |
| Schneider et al.[66] | Caucasian, African-American | 175/520 | N.S. | |
| −7C/T rs25648 | Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| rs833070 C/T (intron 2) | Beeghly-Fadiel et al.[73] | Chinese | 4,419/1,851 | TT genotype was associated with increased risk for breast cancer when compared to CC and CT genotype, OR = 1.26 (1.05–1.52), p = 0.016. |
| SNPs | Authors | Population | Case/control | Susceptibility |
| 936C/T rs3025039 | Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. |
| Krippl et al.[60] | Austrian | 500/500 | T allele had protective effect OR = 0.51, (0.38–0.70), p < 0.001. | |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Eroglu et al.[74] | Turkish | 60/60 | CT was more frequent in patient group (p = 0.001) | |
| Gerger et al.[75] | Austrian | 500/500 | T allele was associated with decreased risk of breast cancer, OR = 0.58 (0.44–0.76), p < 0.001. | |
| Jacobs et al.[65] | American | 504/501 | CC genotype was associated with reduce risk for in situ cancer, OR = 0.59 (0.37–0.93), p = 0.052. | |
| Jakubowska et al.[76] | Polish BRCA1 mutation carriers | 190/319 | CT and TT genotypes were associated with reduced risk of breast cancer, OR = 0.63 (0.41–0.98), p = 0.042. | |
| Jakubowska et al.[77] | Polish | 1015/1073 | N.S. | |
| Jin et al.[51] | Polish German Swedish | 1519/1489 | N.S. | |
| Kataoka et al.[70] | Chinese | 1184/1093 | TT genotype was associated with decreased risk of breast cancer in premenopausal women, OR = 0.45 (0.25–0.79), p = 0.041. | |
| Oliveira et al.[72] | Caucasian, African- American | 235/235 | N.S. | |
| Schneider et al.[66] | Caucasian, African- American | 175/520 | N.S. | |
| Rodrigues et al.[78] | Spanish | 453/461 | CT and TT genotypes had protective effect against breast cancer, OR = 0.67 (0.48–0.92), p (trend) = 0.014 | |
| Zhang et al.[79] | Chinese | 1918/1819 | N.S. | |
| 1612G/A rs10434 | Langsenlehner et al.[68] | Austrian | 804/804 | N.S. |
| SNPs | Authors | Population | Case/control | Aggressiveness |
|---|---|---|---|---|
| −2578C/A rs699947 | Jin et al.[51] | Polish, German, Swedish | 1525/1503 | AA genotype was associated with low grade tumor, p (trend) = 0.04. |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Kidd et al.[67] | Caucasian | 441/− | ER and PR positive patients with CC genotype had higher incidence of recurrent, p (trend) = 0.026 | |
| Etienne-Grimaldi et al.[80] | Caucasian | 137/− | N.S. | |
| −2489C/T rs1005230 | Langsenlehner et al.[68] | Austrian | 804/804 | N.S. |
| −1498C/T rs833061 | Lu et al.[81] | Chinese | 1193/− | CC genotype tended to be associated with decreased OS, HR = 1.5 (0.9–2.5), p (trend) = 0.11. |
| Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. | |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Etienne-Grimaldi et al.[80] | Caucasian | 137/− | N.S. | |
| −1154G/A rs1570360 | Jin et al.[51] | Polish, German | 586/570 | N.S. |
| Smith et al.[71] | English | 263/144 | AG was associated with good prognosis *, OR = 2.63 (1.12–6.20), p = 0.02. GG was associated with negative ER, OR = 0.35 (0.14–0.84), p = 0.02. | |
| Kidd et al.[67] | Caucasian | 441/− | N.S. | |
| Etienne-Grimaldi et al.[80] | Caucasian | 137/− | N.S. | |
| SNPs | Authors | Population | Case/control | Aggressiveness |
| −634G/C rs2010963 | Jin et al.[51] | Swedish | 941/936 | CC genotype was associated with tumor size > 20 mm, OR = 2.20 (1.27–3.82), p = 0.004, and higher histologic grade, p = 0.009. |
| Lu et al.[81] | Chinese | 1193/− | G allele was associated with decreased OS. HR = 1.6 (1.0–2.5) for GG genotype. | |
| Balasubramanian et al.[69] | Caucasian | 500/498 | C allele was associated with maximum size of invasive component, p (trend) = 0.02. | |
| Langsenlehner et al.[68] | Austrian | 804/804 | C allele was associated with small tumor size, p < 0.001. | |
| Oliveira et al.[72] | Caucasian/Afric an-American | 235/235 | N.S. | |
| Kidd et al.[67] | Caucasian | 441/− | N.S. | |
| Etienne-Grimaldi et al.[80] | Caucasian | 137/− | N.S. | |
| Beeghly-Fadiel et al.[73] | Chinese | Stage 1:1193/− Stage 2:5381/− | N.S. | |
| −7C/T rs25648 | Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| 936C/T rs3025039 | Eroglu et al.[74] | Turkish | 60/60 | N.S. |
| Lu et al.[81] | Chinese | 1193/− | N.S. | |
| Wolf et al.[82] | Caucasian | 37/− | Number of T allele was correlated with FDG uptake score, p (Spearman correlation) = 0.032. | |
| Balasubramanian et al.[69] | Caucasian | 500/498 | N.S. | |
| Krippl et al.[60] | Austrian | 500/500 | N.S. | |
| Langsenlehner et al.[68] | Austrian | 804/804 | N.S. | |
| Oliveira et al.[72] | Caucasian/African-American | 235/235 | CC genotype was correlated with increased risk for aggressive breast cancer, OR = 1.76 (1.10–2.90) and negative ER, OR = 0.86 (1.10–3.10). | |
| Knechtel et al.[83] | Caucasian | 432/− | N.S. | |
| Etienne-Grimaldi et al.[80] | Caucasian | 137/− | TT and CT genotypes were associated with longer time to progression when compared to 936 CC genotype (11.5 vs. 9.7 months, p = 0.022). | |
| 1612G/A rs10434 | Langsenlehner et al.[68] | Austrian | 804/804 | N.S. |
| SNPs | Authors | Population | Case/control | Levels of expression | |
|---|---|---|---|---|---|
| Specimen | Results | ||||
| −2578C/A rs699947 | Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. |
| −2489C/T rs1005230 | Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. |
| −1498C/T rs833061 | Balasubramanian et al.[69] | Caucasian | 500/498 | Serum/Tissue | N.S. |
| Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. | |
| −634G/C rs2010963 | Balasubramanian et al.[69] | Caucasian | 500/498 | Serum/Tissue | N.S. |
| Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. | |
| Oliveira et al.[72] | Caucasian, African-American | 235/235 | Serum | N.S. | |
| −7C/T rs25648 | Balasubramanian et al.[69] | Caucasian | 500/498 | Serum/Tissue | N.S. |
| Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. | |
| 936C/T rs3025039 | Balasubramanian et al.[69] | Caucasian | 500/498 | Serum/Tissue | N.S. |
| Krippl et al.[60] | Austrian | −/21 | Plasma | Carriers of T allele tend to had lower VEGFA levels (not reach significant) | |
| Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. | |
| Oliveira et al.[72] | Caucasian, African-American | 235/235 | Serum | N.S. | |
| 1612G/A rs10434 | Langsenlehner et al.[68] | Austrian | −/81 | Plasma | N.S. |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sa-nguanraksa, D.; O-charoenrat, P. The Role of Vascular Endothelial Growth Factor A Polymorphisms in Breast Cancer. Int. J. Mol. Sci. 2012, 13, 14845-14864. https://doi.org/10.3390/ijms131114845
Sa-nguanraksa D, O-charoenrat P. The Role of Vascular Endothelial Growth Factor A Polymorphisms in Breast Cancer. International Journal of Molecular Sciences. 2012; 13(11):14845-14864. https://doi.org/10.3390/ijms131114845
Chicago/Turabian StyleSa-nguanraksa, Doonyapat, and Pornchai O-charoenrat. 2012. "The Role of Vascular Endothelial Growth Factor A Polymorphisms in Breast Cancer" International Journal of Molecular Sciences 13, no. 11: 14845-14864. https://doi.org/10.3390/ijms131114845




