Changes on the Caco-2 Secretome through Differentiation Analyzed by 2-D Differential In-Gel Electrophoresis (DIGE)
Abstract
:1. Introduction
2. Results and Discussion
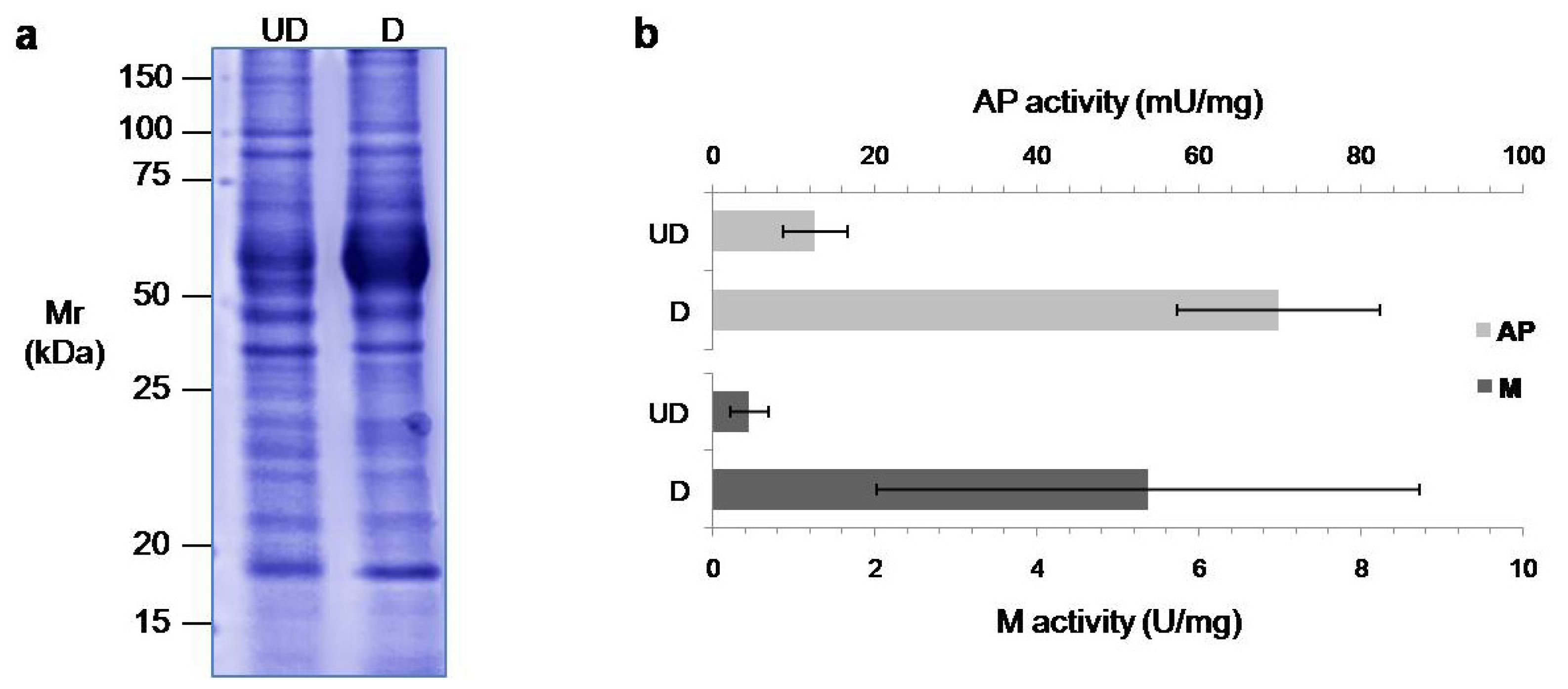
2.1. Differentiation of the Caco-2 Cell Line
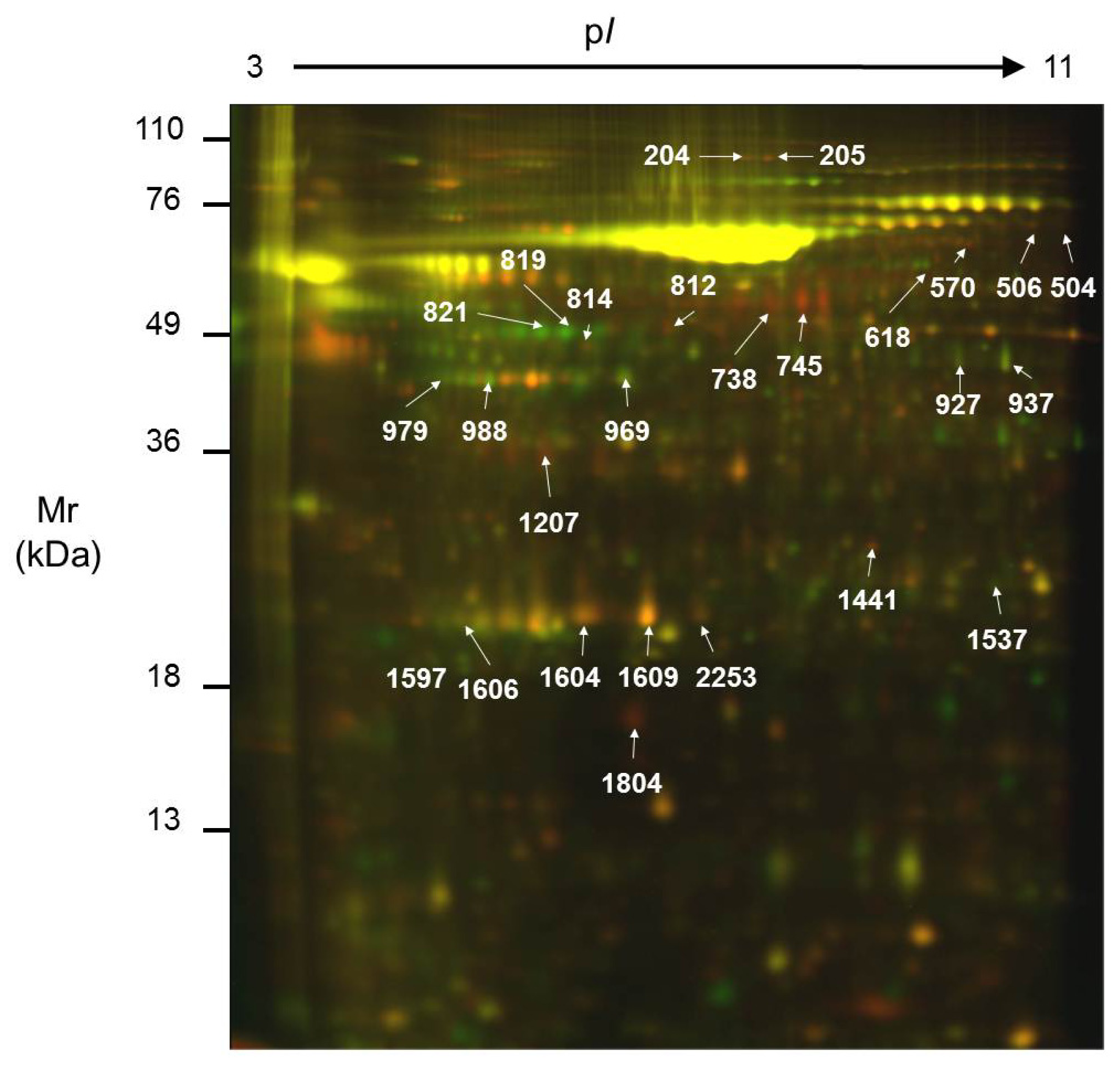
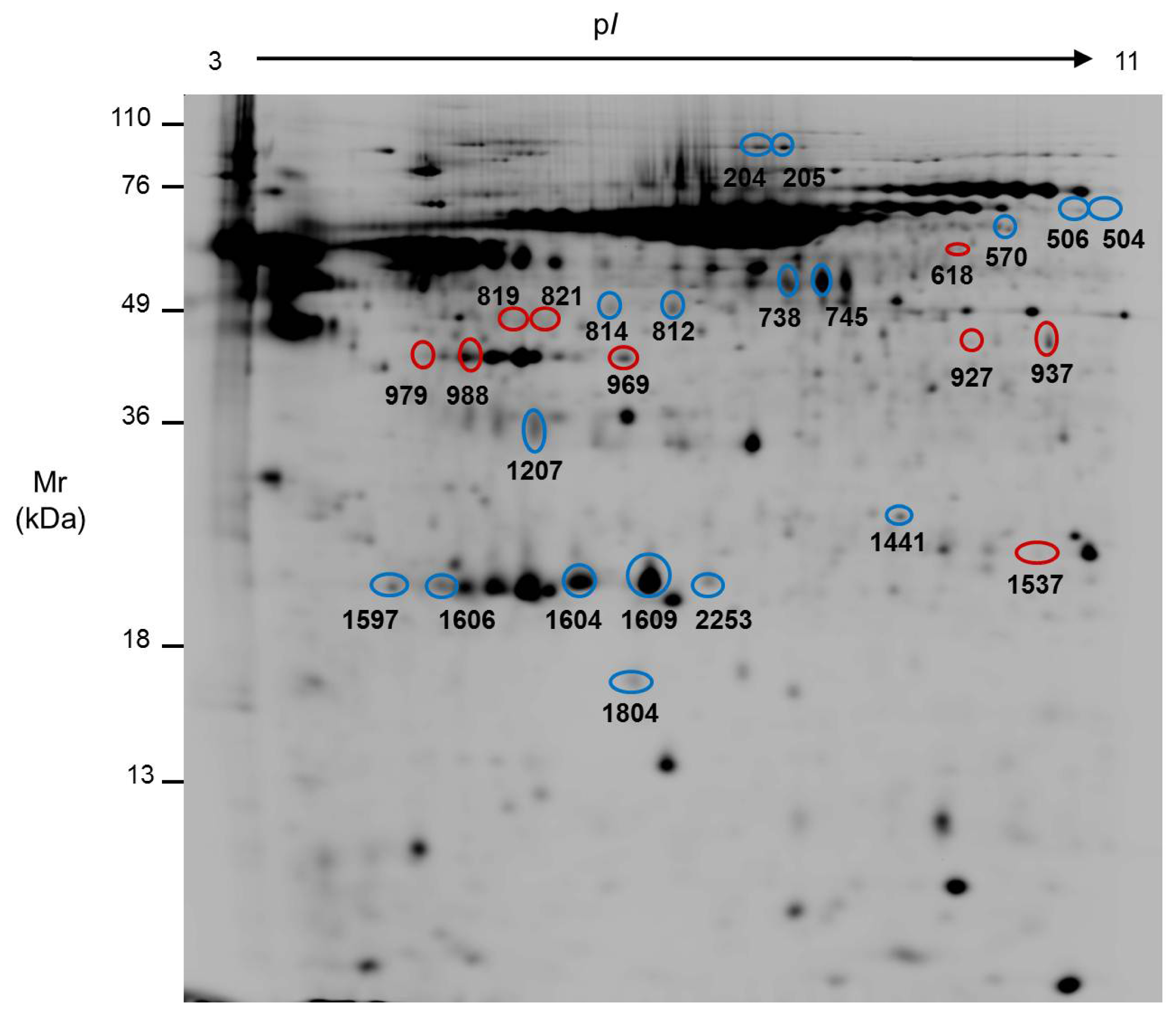
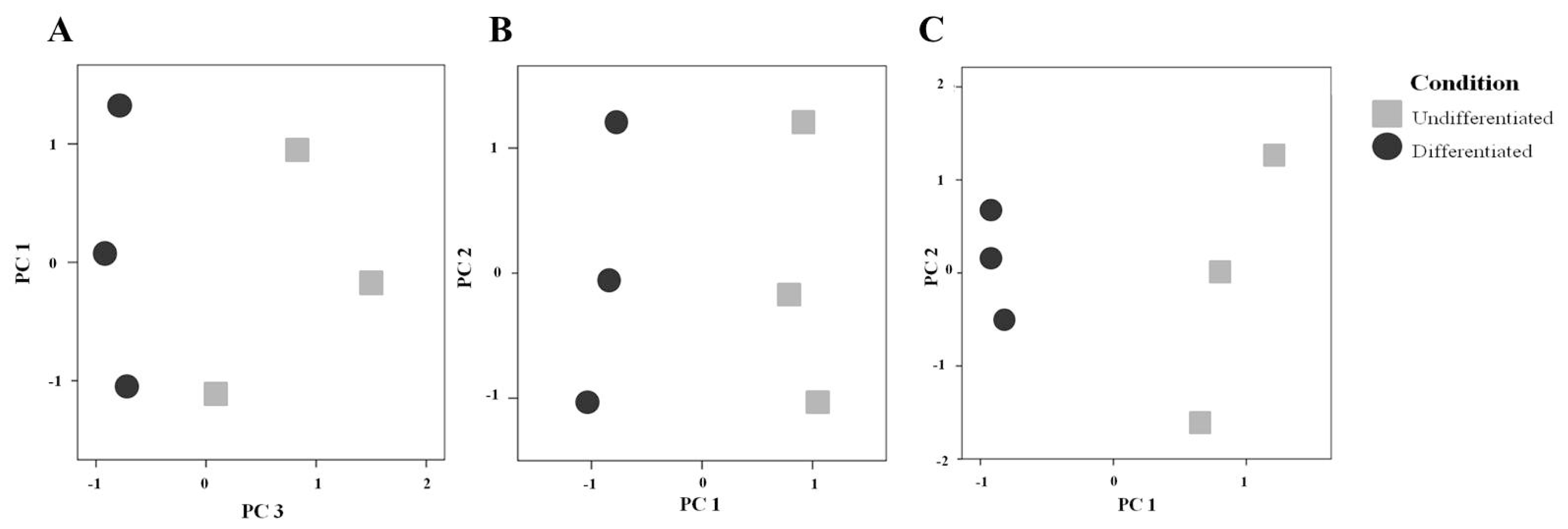
2.2. Comparison of the Secretome of Undifferentiated and Differentiated Caco-2 Cells
3. Experimental Section
3.1. Cell Culture and Enterocytic Differentiation
3.2. Enzymatic Assays
3.3. Secretome Collection and Preparation
3.4. SDS-PAGE
3.5. Differential In-Gel Electrophoresis (DIGE)
3.6. Image Acquisition and Computer Analysis of Electrophoretic Patterns
3.7. Statistical and Computational Methods
3.8. Protein Identification by Mass Spectrometry
4. Conclusions
Supplementary Information
ijms-13-14401-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Schnekenburger, M.; Diederich, M. Epigenetics offer new horizons for colorectal cancer prevention. Curr. Colorectal Cancer Rep 2012, 8, 66–81. [Google Scholar]
- Søreide, K.; Nedrebo, B.S.; Knapp, J.C.; Glomsaker, T.B.; Søreide, J.A.; Korner, H. Evolving molecular classification by genomic and proteomic biomarkers in colorectal cancer: Potential implications for the surgical oncologist. Surg. Oncol 2009, 18, 31–50. [Google Scholar]
- Pinto, M.; Robin-Leon, S.; Appay, M.D.; Kedinger, M.; Triadou, N.; Dussaulx, E.; Lacroix, B.; Simon-Assman, P.; Haffen, K.; Fogh, J.; et al. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 1983, 47, 323–330. [Google Scholar]
- Rousset, M.; Laburthe, M.; Pinto, M.; Chevalier, G.; Rouyer-Fessard, C.; Dussaulx, E.; Trugnan, G.; Boige, N.; Brun, J.L.; Zweibaum, A. Enterocytic differentiation and glucose utilization in the human colon tumor cell line Caco-2: Modulation by forskolin. J. Cell Physiol 1985, 123, 377–385. [Google Scholar]
- Stierum, R.; Gaspari, M.; Dommels, Y.; Ouatas, T.; Pluk, H.; Jespersen, S.; Vogels, J.; Verhoeckx, K.; Groten, J.; van Ommen, B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim. Biophys. Acta 2003, 1650, 73–91. [Google Scholar]
- Buhrke, T.; Lengler, I.; Lampen, A. Analysis of proteomic changes induced upon cellular differentiation of the human intestinal cell line Caco-2. Dev. Growth. Differ 2011, 53, 411–426. [Google Scholar]
- Hauck, W.; Stanners, C.P. Control of carcinoembryonic antigen gene family expression in a differentiating colon carcinoma cell line, Caco-2. Cancer Res 1991, 51, 3526–3533. [Google Scholar]
- Volmer, M.W.; Radacz, Y.; Hahn, S.A.; Klein-Scory, S.; Stühler, K.; Zapatka, M.; Schmiegel, W.; Meyer, H.E.; Schwarte-Waldhoff, I. Tumor suppressor Smad4 mediates downregulation of the anti-adhesive invasion-promoting matricellular protein SPARC: Landscaping activity of Smad4 as revealed by a “secretome” analysis. Proteomics 2004, 4, 1324–1334. [Google Scholar]
- Wu, C.C.; Chien, K.Y.; Tsang, N.M.; Chang, K.P.; Hao, S.P.; Tsao, C.H.; Chang, Y.S.; Yu, J.S. Cancer cell-secreted proteomes as a basis for searching potential tumor markers: Nasopharyngeal carcinoma as a model. Proteomics 2005, 5, 3173–3182. [Google Scholar]
- Wu, C.C.; Chen, H.C.; Chen, S.J.; Liu, H.P.; Hsieh, Y.Y.; Yu, C.J.; Tang, R.; Hsieh, L.L.; Yu, J.S.; Chang, Y.S. Identification of collapsin response mediator protein-2 as a potential marker of colorectal carcinoma by comparative analysis of cancer cell secretomes. Proteomics 2008, 8, 316–332. [Google Scholar]
- Tonge, R.; Shaw, J.; Middleton, B.; Rowlinson, R.; Rayner, S.; Young, J.; Pognan, F.; Hawkins, E.; Currie, I.; Davison, M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 2001, 1, 377–396. [Google Scholar]
- Alban, A.; David, S.O.; Bjorkesten, L.; Andersson, C.; Sloge, E.; Lewis, S.; Currie, I. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003, 3, 36–44. [Google Scholar]
- Unlü, M.; Morgan, M.E.; Minden, J.S. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis 1997, 18, 2071–2077. [Google Scholar]
- Friedman, D.B.; Hill, S.; Keller, J.W.; Merchant, N.B.; Levy, S.E.; Coffey, R.J.; Caprioli, R.M. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 2004, 4, 793–811. [Google Scholar]
- Alfonso, P.; Núñez, A.; Madoz-Gurpide, J.; Lombardia, L.; Sánchez, L.; Casal, J.I. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics 2005, 5, 2602–2611. [Google Scholar]
- Rodríguez-Pineiro, A.M.; Rodríguez-Berrocal, F.J.; Páez de la Cadena, M. Detection of proteins altered in serum of patients with colorectal cancer by differential in-gel electrophoresis. Revista Real Academia Galega de Ciencias 2006, XXV, 5–25. [Google Scholar]
- Seike, M.; Kondo, T.; Fujii, K.; Yamada, T.; Gemma, A.; Kudoh, S.; Hirohashi, S. Proteomic signature of human cancer cells. Proteomics 2004, 4, 2776–2788. [Google Scholar]
- Corbo, C.; Orrù, S.; Gemei, M.; Noto, R.D.; Mirabelli, P.; Imperlini, E.; Ruoppolo, M.; Vecchio, L.D.; Salvatore, F. Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumorigenicity. Proteomics 2012, 12, 2045–2059. [Google Scholar]
- Zeiser, J.J.; Klodmann, J.; Braun, H.P.; Gerhard, R.; Just, I.; Pich, A. Effects of Clostridium difficile Toxin A on the proteome of colonocytes studied by differential 2D electrophoresis. J. Proteomics 2011, 75, 469–479. [Google Scholar]
- Turck, N.; Richert, S.; Gendry, P.; Stutzmann, J.; Kedinger, M.; Leize, E.; Simon-Assmann, P.; Van Dorsselaer, A.; Launay, J.F. Proteomic analysis of nuclear proteins from proliferative and differentiated human colonic intestinal epithelial cells. Proteomics 2004, 4, 93–105. [Google Scholar]
- Pshezhetsky, A.V.; Fedjaev, M.; Ashmarina, L.; Mazur, A.; Budman, L.; Sinnett, D.; Labuda, D.; Beaulieu, J.F.; Ménard, D.; Nifant’ev, I.; et al. Subcellular proteomics of cell differentiation: Quantitative analysis of the plasma membrane proteome of Caco-2 cells. Proteomics 2007, 7, 2201–2215. [Google Scholar]
- Mariadason, J.M.; Arango, D.; Corner, G.A.; Arañes, M.J.; Hotchkiss, K.A.; Yang, W.; Augenlicht, L.H. A gene expression profile that defines colon cell maturation in vitro. Cancer Res 2002, 62, 4791–4804. [Google Scholar]
- Tremblay, E.; Auclair, J.; Delvin, E.; Levy, E.; Ménard, D.; Pshezhetsky, A.V.; Rivard, N.; Seidman, E.G.; Sinnett, D.; Vachon, P.H.; et al. Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: A comparison with the Caco-2 cell model. J. Cell. Biochem 2006, 99, 1175–1186. [Google Scholar]
- Volmer, M.W.; Stühler, K.; Zapatka, M.; Schöneck, A.; Klein-Scory, S.; Schmiegel, W.; Meyer, H.E.; Schwarte-Waldhoff, I. Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics 2005, 5, 2587–2601. [Google Scholar]
- Xue, H.; Lü, B.; Zhang, J.; Wu, M.; Huang, Q.; Wu, Q.; Sheng, H.; Wu, D.; Hu, J.; Lai, M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res 2010, 9, 545–555. [Google Scholar]
- Mbeunkui, F.; Metge, B.J.; Shevde, L.A.; Pannell, L.K. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J. Proteome Res 2007, 6, 2993–3002. [Google Scholar]
- Ji, H.; Greening, D.W.; Kapp, E.A.; Moritz, R.L.; Simpson, R.J. Secretome-based proteomics reveals sulindac-modulated proteins released from colon cancer cells. Proteomics Clin. Appl 2009, 3, 433–451. [Google Scholar]
- Yenofsky, R.; Cereghini, S.; Krowczynska, A.; Brawerman, G. Regulation of mRNA utilization in mouse erythroleukemia cells induced to differentiate by exposure to dimethyl sulfoxide. Mol. Cell. Biol 1983, 3, 1197–1203. [Google Scholar]
- Nagano-Ito, M.; Ichikawa, S. Biological effects of Mammalian translationally controlled tumor protein (TCTP) on cell death, proliferation, and tumorigenesis. Biochem. Res. Int 2012, 2012, 204960–204966. [Google Scholar]
- MacDonald, S.M.; Paznekas, W.A.; Jabs, E.W. Chromosomal localization of tumor protein, translationally-controlled 1 (TPT1) encoding the human histamine releasing factor (HRF) to 13q12→q14. Cytogenet. Cell. Genet 1999, 84, 128–129. [Google Scholar]
- Arcuri, F.; Papa, S.; Carducci, A.; Romagnoli, R.; Liberatori, S.; Riparbelli, M.G.; Sanchez, J.C.; Tosi, P.; del Vecchio, M.T. Translationally controlled tumor protein (TCTP) in the human prostate and prostate cancer cells: Expression, distribution, and calcium binding activity. Prostate 2004, 60, 130–140. [Google Scholar]
- Li, F.; Zhang, D.; Fujise, K. Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem 2001, 276, 47542–47549. [Google Scholar]
- Teshima, S.; Rokutan, K.; Nikawa, T.; Kishi, K. Macrophage colony-stimulating factor stimulates synthesis and secretion of a mouse homolog of a human IgE-dependent histamine-releasing factor by macrophages in vitro and in vivo. J. Immunol 1998, 161, 6356–6366. [Google Scholar]
- Amzallag, N.; Passer, B.J.; Allanic, D.; Segura, E.; Théry, C.; Goud, B.; Amson, R.; Telerman, A. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. Biol. Chem 2004, 279, 46104–46112. [Google Scholar]
- Chung, S.; Kim, M.; Choi, W.J.; Chung, J.K.; Lee, K. Expression of translationally controlled tumor protein mRNA in human colon cancer. Cancer Lett 2000, 156, 185–190. [Google Scholar]
- Ma, Q.; Geng, Y.; Xu, W.; Wu, Y.; He, F.; Shu, W.; Huang, M.; Du, H.; Li, M. The role of translationally controlled tumor protein in tumor growth and metastasis of colon adenocarcinoma cells. J. Proteome Res 2010, 9, 40–49. [Google Scholar]
- Skonier, J.; Neubauer, M.; Madisen, L.; Bennett, K.; Plowman, G.D.; Purchio, A.F. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol 1992, 11, 511–522. [Google Scholar]
- Skonier, J.; Bennett, K.; Rothwell, V.; Kosowski, S.; Plowman, G.; Wallace, P.; Edelhoff, S.; Disteche, C.; Neubauer, M.; Marquardt, H. Beta ig-h3: A transforming growth factor-beta-responsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA Cell Biol 1994, 13, 571–584. [Google Scholar]
- Thapa, N.; Lee, B.H.; Kim, I.S. TGFBIp/betaig-h3 protein: A versatile matrix molecule induced by TGF-beta. Int. J. Biochem. Cell Biol 2007, 39, 2183–2194. [Google Scholar]
- Kitahara, O.; Furukawa, Y.; Tanaka, T.; Kihara, C.; Ono, K.; Yanagawa, R.; Nita, M.E.; Takagi, T.; Nakamura, Y.; Tsunoda, T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res 2001, 61, 3544–3559. [Google Scholar]
- Buckhaults, P.; Rago, C.; St Croix, B.; Romans, K.E.; Saha, S.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res 2001, 61, 6996–7001. [Google Scholar]
- Roessler, M.; Rollinger, W.; Palme, S.; Hagmann, M.L.; Berndt, P.; Engel, A.M.; Schneidinger, B.; Pfeffer, M.; Andres, H.; Karl, J.; et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin. Cancer Res 2005, 11, 6550–6557. [Google Scholar]
- Ma, C.; Rong, Y.; Radiloff, D.R.; Datto, M.B.; Centeno, B.; Bao, S.; Cheng, A.W.; Lin, F.; Jiang, S.; Yeatman, T.J.; et al. Extracellular matrix protein betaig-h3/TGFBI promotes metastasis of colon cancer by enhancing cell extravasation. Genes Dev 2008, 22, 308–321. [Google Scholar]
- Chang, Y.T.; Wu, C.C.; Shyr, Y.M.; Chen, T.C.; Hwang, T.L.; Yeh, T.S.; Chang, K.P.; Liu, H.P.; Liu, Y.L.; Tsai, M.H.; et al. Secretome-based identification of ULBP2 as a novel serum marker for pancreatic cancer detection. PLoS One 2011, 6, e20029. [Google Scholar]
- Naureckiene, S.; Sleat, D.E.; Lackland, H.; Fensom, A.; Vanier, M.T.; Wattiaux, R.; Jadot, M.; Lobel, P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science 2000, 290, 2298–2301. [Google Scholar]
- Larsen, L.B.; Ravn, P.; Boisen, A.; Berglund, L.; Petersen, T.E. Primary structure of EPV20, a secretory glycoprotein containing a previously uncharacterized type of domain. Eur. J. Biochem 1997, 243, 437–441. [Google Scholar]
- Kirchhoff, C.; Osterhoff, C.; Young, L. Molecular cloning and characterization of HE1, a major secretory protein of the human epididymis. Biol. Reprod 1996, 54, 847–856. [Google Scholar]
- Klein, A.; Amigo, L.; Retamal, M.J.; Morales, M.G.; Miquel, J.F.; Rigotti, A.; Zanlungo, S. NPC2 is expressed in human and murine liver and secreted into bile: Potential implications for body cholesterol homeostasis. Hepatology 2006, 43, 126–133. [Google Scholar]
- Storch, J.; Xu, Z. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim. Biophys. Acta 2009, 1791, 671–678. [Google Scholar]
- Heo, K.; Jariwala, U.; Woo, J.; Zhan, Y.; Burke, K.A.; Zhu, L.; Anderson, W.F.; Zhao, Y. Involvement of Niemann-Pick type C2 protein in hematopoiesis regulation. Stem Cells 2006, 24, 1549–1555. [Google Scholar]
- Schrantz, N.; Sagiv, Y.; Liu, Y.; Savage, P.B.; Bendelac, A.; Teyton, L. The Niemann-Pick type C2 protein loads isoglobotrihexosylceramide onto CD1d molecules and contributes to the thymic selection of NKT cells. J. Exp. Med 2007, 204, 841–852. [Google Scholar]
- Csepeggi, C.; Jiang, M.; Frolov, A. Somatic cell plasticity and Niemann-Pick type C2 protein: Adipocyte differentiation and function. J. Biol. Chem 2010, 285, 30347–30354. [Google Scholar]
- Csepeggi, C.; Jiang, M.; Kojima, F.; Crofford, L.J.; Frolov, A. Somatic cell plasticity and Niemann-Pick type C2protein. J. Biol. Chem 2011, 286, 2078–2087. [Google Scholar]
- McDonald, S.L.; Edington, H.D.; Kirkwood, J.M.; Becker, D. Expression analysis of genes identified by molecular profiling of VGP melanomas and MGP melanoma-positive lymph nodes. Cancer Biol. Ther 2004, 3, 110–120. [Google Scholar]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar]
- Pang, B.; Zhang, H.; Wang, J.; Chen, W.Z.; Li, S.H.; Shi, Q.G.; Liang, R.X.; Xie, B.X.; Wu, R.Q.; Qian, X.L.; et al. Ubiquitous mitochondrial creatine kinase is overexpressed in the conditioned medium and the extract of LNCaP lineaged androgen independent cell lines and facilitates prostate cancer progression. Prostate 2009, 69, 1176–1187. [Google Scholar]
- Engstrom, L. Alkaline and acid phosphatase. In Preparative Centrifugation. A Practical Approach; Rickwood, D., Hames, B.D., Eds.; IRL Press: Oxford, UK, 1992; p. 375. [Google Scholar]
- Dahlqvist, A. Assay of intestinal disaccharidases. Scand. J. Clin. Lab. Invest 1984, 44, 169–172. [Google Scholar]
- Trinder, P. Determination of blood glucose using an oxidase-peroxidase system with a non-carcinogenic chromogen. J. Clin. Pathol 1969, 22, 158–161. [Google Scholar]
- Trinder, P. Determination of blood glucose using 4-amino phenazone as oxygen acceptor. J. Clin. Pathol 1969, 22, 246. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Beyer, E.; Ivleva, T.; Artykova, G.; Wiederschain, G. Change of isoforms’ spectra of alpha-l-fucosidase from human skin fibroblasts in intracellular storage of non hydrolyzable substances. Biochim. Biophys. Acta 1995, 1270, 7–11. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Ramagli, L.S.; Rodriguez, V.S. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 1985, 6, 559–563. [Google Scholar]
- Rodríguez-Piñeiro, A.M.; Rodríguez-Berrocal, F.J.; Páez de la Cadena, M. Improvements in the search for potential biomarkers by proteomics: Application of principal component and discriminant analyses for two-dimensional maps evaluation. J. Chromatogr. B 2007, 849, 251–260. [Google Scholar]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel 2004, 17, 349–356. [Google Scholar]
| Gel | No. Spots Detected | No. Spots Matched | * Label | Sample |
|---|---|---|---|---|
| Gel 1 | 2254 | 2254 | Cy2 | Standard |
| Cy5 | UD1 | |||
| Cy3 | D3 | |||
| Gel 2 | 1632 | 1164 | Cy2 | Standard |
| Cy5 | UD2 | |||
| Cy3 | D1 | |||
| Gel 3 | 2023 | 1463 | Cy2 | Standard |
| Cy5 | D2 | |||
| Cy3 | UD3 | |||
| Spot No. | ID | Name | pI | Mr (kDa) | Int. UD (mean ± SD) | Int. D (mean ± SD) | Ratio | Function | Localization | Method | Protein Score | Threshold | Coverage (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 204 | Q14697 | Glucosidase II subunit alpha | 5.7 | 107.2 | 184.36 ± 113.39 | 61.32 ± 17.73 | 3.0 | N-glycoprotein metabolism | ER; Pred. sec. | PMF | 249 | 66 | 34 |
| 205 | Q14697 | Glucosidase II subunit alpha | 5.7 | 107.2 | 126.37 ± 61.35 | 32.18 ± 7.26 | 3.9 | N-glycoprotein metabolism | ER; Pred. sec. | PMF | 275 | 66 | 37 |
| 504 | Q15582 | Transforming growth factor-betainduced protein ig-h3 | 7.7 | 74.7 | 30.31 ± 3.72 | 15.50 ± 2.20 | 2.0 | Cellular adhesion | Secreted | SQ | 107 | 66 | 21 |
| 506 | Q15582 | Transforming growth factor-betainduced protein ig-h3 | 7.7 | 74.7 | 65.28 ± 17.57 | 31.04 ± 3.54 | 2.1 | Cellular adhesion | Secreted | PMF | 77 | 66 | 25 |
| 570 | Q15582 | Transforming growth factor-betainduced protein ig-h3 | 7.7 | 74.7 | 60.17 ± 10.52 | 22.14 ± 6.58 | 2.7 | Cellular adhesion | Secreted | SQ | 211 | 56 | 29 |
| 618 | P23141 | Liver carboxylesterase1 | 5.9 | 56.5 | 24.86 ± 10.16 | 58.72 ± 19.93 | 0.4 | Detoxification | ER; Pred. sec. | PMF | 91 | 66 | 34 |
| 738 | P10619 | Cathepsin A | 6.0 | 51.9 | 959.27 ± 685.01 | 229.14 ± 142.67 | 4.2 | Carboxipeptidase | Lysosome; Pred. sec. | SQ | 104 | 66 | 16 |
| 745 | P06576 | ATP synthase subunit beta | 5.3 | 56.5 | 2445.77 ± 2596.37 | 360.55 ± 231.07 | 6.8 | ATP synthesis | Mitochondria; Pred. sec. | SQ | 115 | 56 | 27 |
| 814 | P02679 | Fibrinogen gamma chain | 5.5 | 46.8 | 141.99 ± 34.59 | 54.12 ± 23.49 | 2.6 | Hemostasis/acute phase response | Secreted | SQ | 97 | 66 | 14 |
| 927 | Q6FHQ6 | NADPdependent isocitrate dehydrogenase | 6.5 | 46.9 | 80.32 ± 6.07 | 201.39 ± 91.20 | 0.4 | Carbohydrate metabolism | Cytosol; Pred. Sec. | SQ | 224 | 66 | 23 |
| 937 | Q6FHQ6 | NADPdependent isocitrate dehydrogenase | 6.5 | 46.9 | 242.59 ± 91.94 | 569.91 ± 255.38 | 0.4 | Carbohydrate metabolism | Cytosol; Pred. Sec. | PMF | 205 | 66 | 50 |
| 969 | P12277 | Creatine kinase B | 5.3 | 42.9 | 253.51 ± 103.76 | 918.55 ± 495.43 | 0.3 | Energy metabolism | Cytosol | PMF | 252 | 66 | 56 |
| 979 | P06727 | Apolipoprotein A-IV | 5.3 | 45.4 | 62.39 ± 32.05 | 321.76 ± 235.62 | 0.2 | Lipid transport | Secreted | PMF | 215 | 66 | 49 |
| 988 | P06727 | Apolipoprotein A-IV | 5.3 | 45.4 | 206.98 ± 52.20 | 723.07 ± 303.91 | 0.3 | Lipid transport | Secreted | SQ | 130 | 66 | 34 |
| 1207 | P02649 | Apolipoprotein E precursor | 5.7 | 36.3 | 354.49 ± 32.66 | 167.01 ± 85.65 | 2.1 | Lipid transport | Secreted | PMF | 215 | 66 | 60 |
| 1441 | P78417 | Glutathione S-transferase omega-1 | 6.2 | 27.8 | 103.79 ± 13.36 | 65.54 ± 33.38 | 1.6 | Glutathione Transference | Cytosol | SQ | 89 | 56 | 17 |
| 1537 | P60174 | Triose phosphate isomerase | 6.5 | 26.8 | 63.43 ± 22.22 | 162.21 ± 95.92 | 0.4 | Carbohydrate metabolism | Cytosol; Pred. sec. | PMF | 141 | 66 | 60 |
| 1597 | Q5W0H4 | Translationallycontrolled tumor protein | 5.1 | 22.8 | 152.49 ± 38.04 | 45.49 ± 8.91 | 3.4 | Calcium binding and microtubule stabilization | Cytosol; Pred. sec. | SQ | 82 | 66 | 35 |
| 1604 | P02647 | Apolipoprotein A-I | 5.5 | 28.9 | 1935.86 ± 793.28 | 943.66 ± 494.74 | 2.1 | Lipid transport | Secreted | PMF | 191 | 66 | 67 |
| 1609 | P02647 | Apolipoprotein A-I | 5.5 | 28.9 | 4306.10 ± 2751.70 | 1567.04 ± 913.34 | 2.8 | Lipid transport | Secreted | PMF | 270 | 66 | 81 |
| 1804 | P61916 | Niemann-Pick disease type C2 protein | 5.3 | 16.6 | 437.13 ± 260.61 | 68.25 ± 49.70 | 6.4 | Cholesterol binding | Secreted | SQ | 101 | 66 | 30 |
| 2253 | P02647 | Apolipoprotein A-I | 5.5 | 28.9 | 302.18 ± 178.79 | 118.77 ± 34.88 | 2.5 | Lipid transport | Secreted | PMF | 68 | 66 | 37 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
García-Lorenzo, A.; Rodríguez-Piñeiro, A.M.; Rodríguez-Berrocal, F.J.; Cadena, M.P.d.l.; Martínez-Zorzano, V.S. Changes on the Caco-2 Secretome through Differentiation Analyzed by 2-D Differential In-Gel Electrophoresis (DIGE). Int. J. Mol. Sci. 2012, 13, 14401-14420. https://doi.org/10.3390/ijms131114401
García-Lorenzo A, Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, Cadena MPdl, Martínez-Zorzano VS. Changes on the Caco-2 Secretome through Differentiation Analyzed by 2-D Differential In-Gel Electrophoresis (DIGE). International Journal of Molecular Sciences. 2012; 13(11):14401-14420. https://doi.org/10.3390/ijms131114401
Chicago/Turabian StyleGarcía-Lorenzo, Andrés, Ana M. Rodríguez-Piñeiro, Francisco J. Rodríguez-Berrocal, María Páez de la Cadena, and Vicenta S. Martínez-Zorzano. 2012. "Changes on the Caco-2 Secretome through Differentiation Analyzed by 2-D Differential In-Gel Electrophoresis (DIGE)" International Journal of Molecular Sciences 13, no. 11: 14401-14420. https://doi.org/10.3390/ijms131114401