Biochip-Based Detection of KRAS Mutation in Non-Small Cell Lung Cancer
Abstract
:1. Introduction
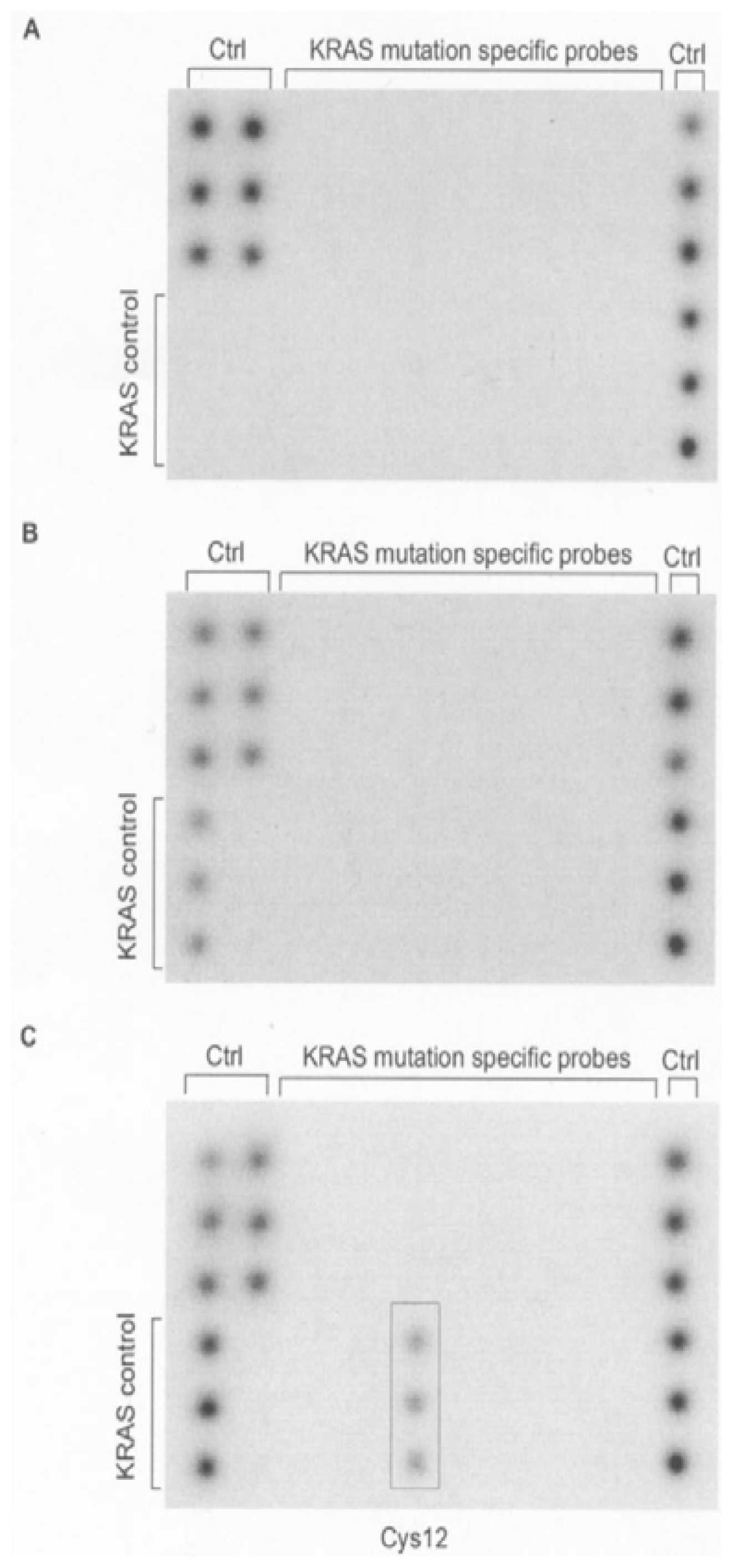
2. Results and Discussion
3. Experimental Section
3.1. Tissue Samples and DNA Isolation
3.2. Mutant-Enriched PCR and Biochip Hybridization
3.3. Dideoxy Sequencing
4. Conclusions
- Conflict of InterestG.K. is an employee of ViennaLab Diagnostics GmbH.
References
- Blons, H.; Pallier, K.; Le Corre, D.; Danel, C.; Tremblay-Gravel, M.; Houdayer, C.; Fabre-Guillevin, E.; Riquet, M.; Dessen, P.; Laurent-Puig, P. Genome wide SNP comparative analysis between EGFR and KRAS mutated NSCLC and characterization of two models of oncogenic cooperation in non-small cell lung carcinoma. BMC Med. Genomics 2008, 1, 25. [Google Scholar]
- Rusch, V.; Baselga, J.; Cordon-Cardo, C.; Orazem, J.; Zaman, M.; Hoda, S.; McIntosh, J.; Kurie, J.; Dmitrovsky, E. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res 1993, 53, 2379–2385. [Google Scholar]
- Adjei, A.A. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst 2001, 93, 1062–1074. [Google Scholar]
- Langer, C.J. Roles of EGFR and KRAS Mutations in the Treatment of Pateintes with Non-Small-Cell Lung Cancer. Pharm. Ther 2011, 36, 263–279. [Google Scholar]
- Keohavong, P.; Mady, H.H.; Gao, W.M.; Siegfried, J.M.; Luketich, J.D.; Melhem, M.F. Topographic analysis of K-ras mutations in histologically normal lung tissues and tumours of lung cancer patients. Br. J. Cancer 2001, 85, 235–241. [Google Scholar]
- Herbst, R.S.; Sandler, A. Bevacizumab and erlotinib: a promising new approach to the treatment of advanced NSCLC. Oncologist 2008, 13, 1166–1176. [Google Scholar]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with the sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar]
- Pao, W.; Wang, T.Y.; Riely, G.J.; Miller, V.A.; Pan, Q.; Ladanyi, M.; Zakowski, M.F.; Hellan, R.T.; Kris, M.-G.; Varmus, H.E. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med 2005, 2, e17. [Google Scholar]
- Fabjani, G.; Kriegshaeuser, G.; Schuetz, A.; Prix, L.; Zeillinger, R. Biochip for K-ras mutation screening in ovarian cancer. Clin. Chem 2005, 51, 784–787. [Google Scholar]
- Auner, V.; Kriegshäuser, G.; Tong, D.; Horvat, R.; Reinthaller, A.; Mustea, A.; Zeillinger, R. KRAS mutation analysis in ovarian samples using a high sensitivity biochip assay. BMC Cancer 2009, 9, e111. [Google Scholar]
- Kriegshäuser, G.; Auner, V.; Schuster, E.; Holzer, B.; Oberkanins, C.; Horvat, R.; Speiser, P.; Zeillinger, R. KRAS mutation analysis in genomic DNA isolated from formalin-fixed paraffin-embedded ovarian tissue: evaluation of a strip-based reverse-hybridisation assay. J. Clin. Pathol 2011, 64, 252–256. [Google Scholar]
- Keohavong, P.; DeMichele, M.A.; Melacrinos, A.C.; Landreneau, R.J.; Weyant, R.J.; Siegfried, J.M. Detection of K-ras mutations in lung carcinomas: Relationship to prognosis. Clin. Cancer Res 1996, 2, 411–418. [Google Scholar]
- Do, H.; Krypuy, M.; Mitchell, P.L.; Fox, S.B.; Dobrovic, A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer 2008, 8, e142. [Google Scholar]
- van Eijk, R.; Licht, J.; Schrumpf, M.; Yazdi, M.T.; Ruano, D.; Forte, G.I.; Nederlof, P.M.; Veselic, M.; Rabe, K.F.; Annema, J.T.; et al. Rapid KRAS, EGFR, BRAF and PIK3CA Mutation Analysis of Fine Needle Aspirates from Non-Small-Cell Lung Cancer Using Allele-Speific qPCR. PLoS One 2011, 6, e17791. [Google Scholar]
- Kotoula, V.; Charalambous, E.; Biesmans, B.; Malousi, A.; Vrettou, E.; Fountzilas, G.; Karkavelas, G. Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics. PLoS One 2009, 4, e7746. [Google Scholar]
- Sarasqueta, A.F.; Moerland, E.; de Bruyne, H.; de raaf, H.; Vrancken, T.; van Lijnschoten, G.; van den Brule, A.J.C. SnaPshot and StripAssay as Valuable Alternatives to Direct Sequencing for KRAS Mutation Detection in Colon Cancer Routine Diagnostics. J. Mol. Diagn 2011, 13, 199–205. [Google Scholar]
- Krypuy, M.; Newnham, G.M.; Thomas, D.M.; Conron, M.; Dobrovic, A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 2006, 6, e295. [Google Scholar]
- Ogino, S.; Kawasaki, T.; Brahmandam, M.; Yan, L.; Cantor, M.; Namgyal, C.; Mino-Kenudson, M.; Lauwers, G.Y.; Loda, M.; Fuchs, C.S. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J. Mol. Diagn 2005, 7, 413–421. [Google Scholar]
- Tol, J.; Dijkstra, J.R.; Vink-Borger, M.E.; Nagtegaal, I.D.; Punt, C.J.; van Krieken, J.H.; Ligtenberg, M.J. High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J. Cell. Mol. Med 2010, 14, 2122–2131. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Prix, L.; Uciechowski, P.; Böckmann, B.; Giesing, M.; Schuetz, A.J. Diagnostic biochip array for fast and sensitive detection of K-ras mutations in stool. Clin. Chem 2002, 48, 428–435. [Google Scholar]
| Characteristic | n | Mutated, n (%) | |
|---|---|---|---|
| Total | 81 | 17 (21) | |
| Gender | Male | 59 | 11 (19) |
| Female | 22 | 6 (27) | |
| Pathology | Squamous cell carcinoma | 30 | 1 (3) |
| Adenocarcinoma | 48 | 16 (33) | |
| Large cell carcinoma | 3 | 0 (0) | |
| Differentiation | Grade 1 | 3 | 0 (0) |
| Grade 2 | 53 | 12 (23) | |
| Grade 3 | 20 | 3 (15) | |
| Unknown | 5 | 2 (40) | |
| Disease stage | IA | 24 | 8 (33) |
| IB | 15 | 2 (13) | |
| IIA | 11 | 4 (36) | |
| IIB | 14 | 0 (0) | |
| IIIA | 15 | 3 (20) | |
| IIIB | 2 | 0 (0) | |
| Pathologic tumor status | pT1 | 32 | 11 (34) |
| pT2 | 40 | 5 (12) | |
| pT3 | 7 | 1 (14) | |
| pT4 | 2 | 0 (0) | |
| Pathologic lymph node status | pN0 | 45 | 10 (22) |
| pN1 | 22 | 5 (23) | |
| pN2 | 14 | 2 (14) | |
| Mutation | Amino acid | n | % |
|---|---|---|---|
| GGT→GAT | Gly12→Asp12 | 6 | 35 |
| GGT→TGT | Gly12→Cys12 | 5 | 29 |
| GGT→GTT | Gly12→Val12 | 3 | 18 |
| GGT→GCT | Gly12→Ala12 | 2 | 12 |
| GGT→AGT | Gly12→Ser12 | 1 | 6 |
| GGT→CGT | Gly12→Arg12 | 0 | 0 |
| GGT→ATT | Gly12→Ile12 | 0 | 0 |
| GGT→CTT | Gly12→Leu12 | 0 | 0 |
| GGC→GAC | Gly13→Asp13 | 0 | 0 |
| GGC→TGC | Gly13→Cys13 | 0 | 0 |
| Total | 17 | 100 | |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kriegshäuser, G.; Fabjani, G.; Ziegler, B.; Zöchbauer-Müller, S.; End, A.; Zeillinger, R. Biochip-Based Detection of KRAS Mutation in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2011, 12, 8530-8538. https://doi.org/10.3390/ijms12128530
Kriegshäuser G, Fabjani G, Ziegler B, Zöchbauer-Müller S, End A, Zeillinger R. Biochip-Based Detection of KRAS Mutation in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences. 2011; 12(12):8530-8538. https://doi.org/10.3390/ijms12128530
Chicago/Turabian StyleKriegshäuser, Gernot, Gerhild Fabjani, Barbara Ziegler, Sabine Zöchbauer-Müller, Adelheid End, and Robert Zeillinger. 2011. "Biochip-Based Detection of KRAS Mutation in Non-Small Cell Lung Cancer" International Journal of Molecular Sciences 12, no. 12: 8530-8538. https://doi.org/10.3390/ijms12128530





