Biochemical Properties and Potential Applications of Recombinant Leucine Aminopeptidase from Bacillus kaustophilus CCRC 11223
Abstract
:1. Introduction
2. Results and Discussion
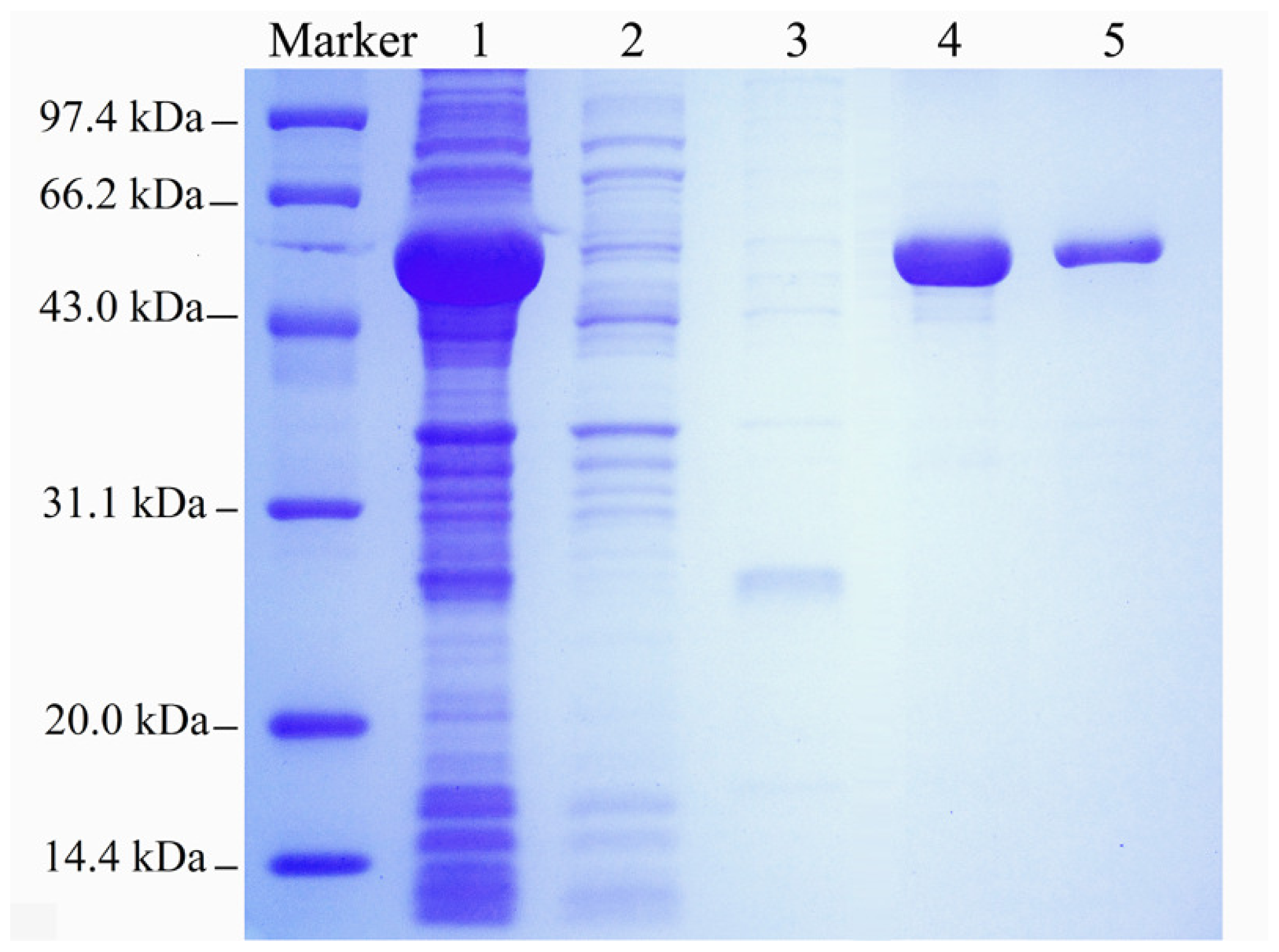
2.1. Expression and Purification of the Recombinant Enzyme
2.2. Biochemical Properties and Conformation Changes of the BkLAP under Different Conditions
2.2.1. Effect of pH on the Enzymatic Activity and Secondary Structure of BkLAP
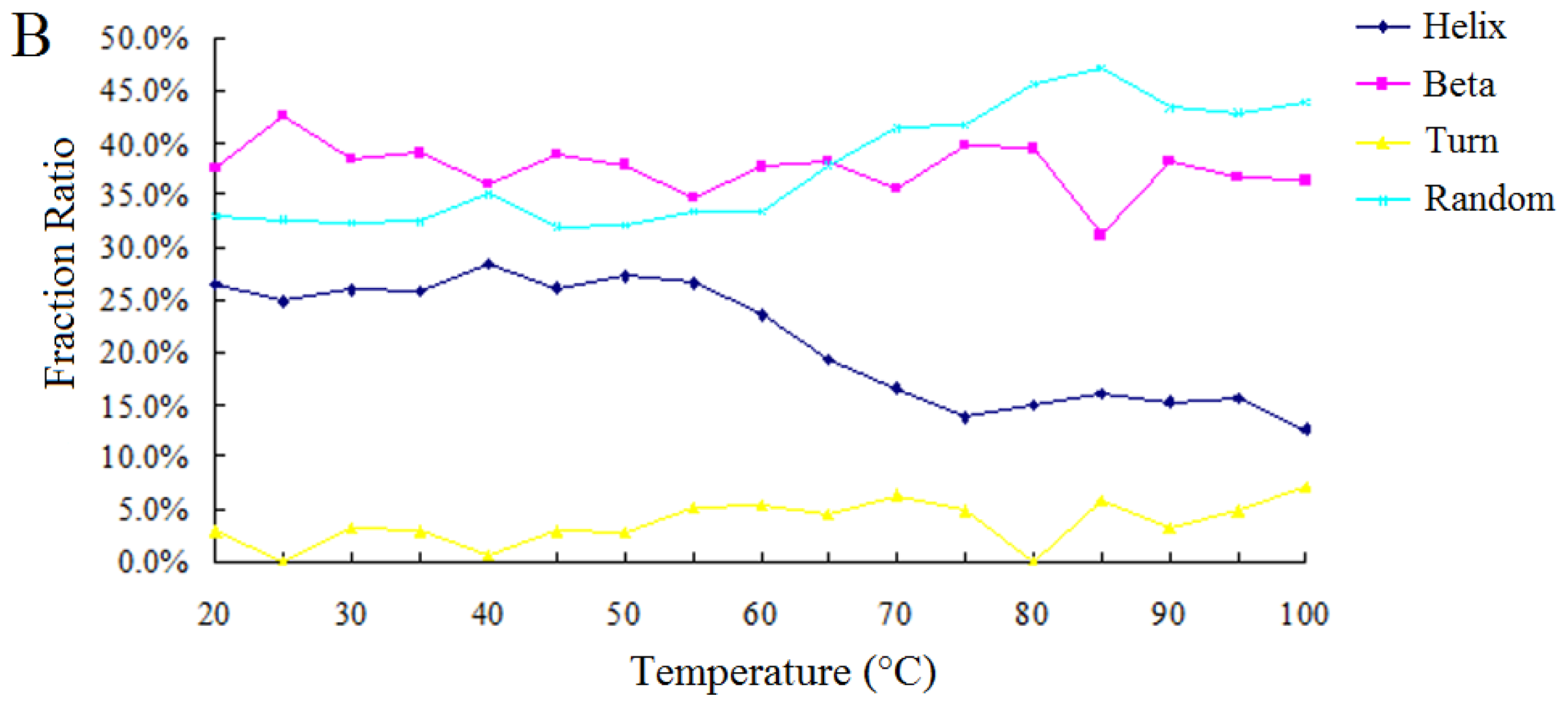
2.2.2. Effect of Temperature on the Enzymatic Activity and the Secondary Structure of BkLAP
2.2.3. Effect of Various Divalent Cations on the Enzymatic Activity and Secondary Structure of BkLAP
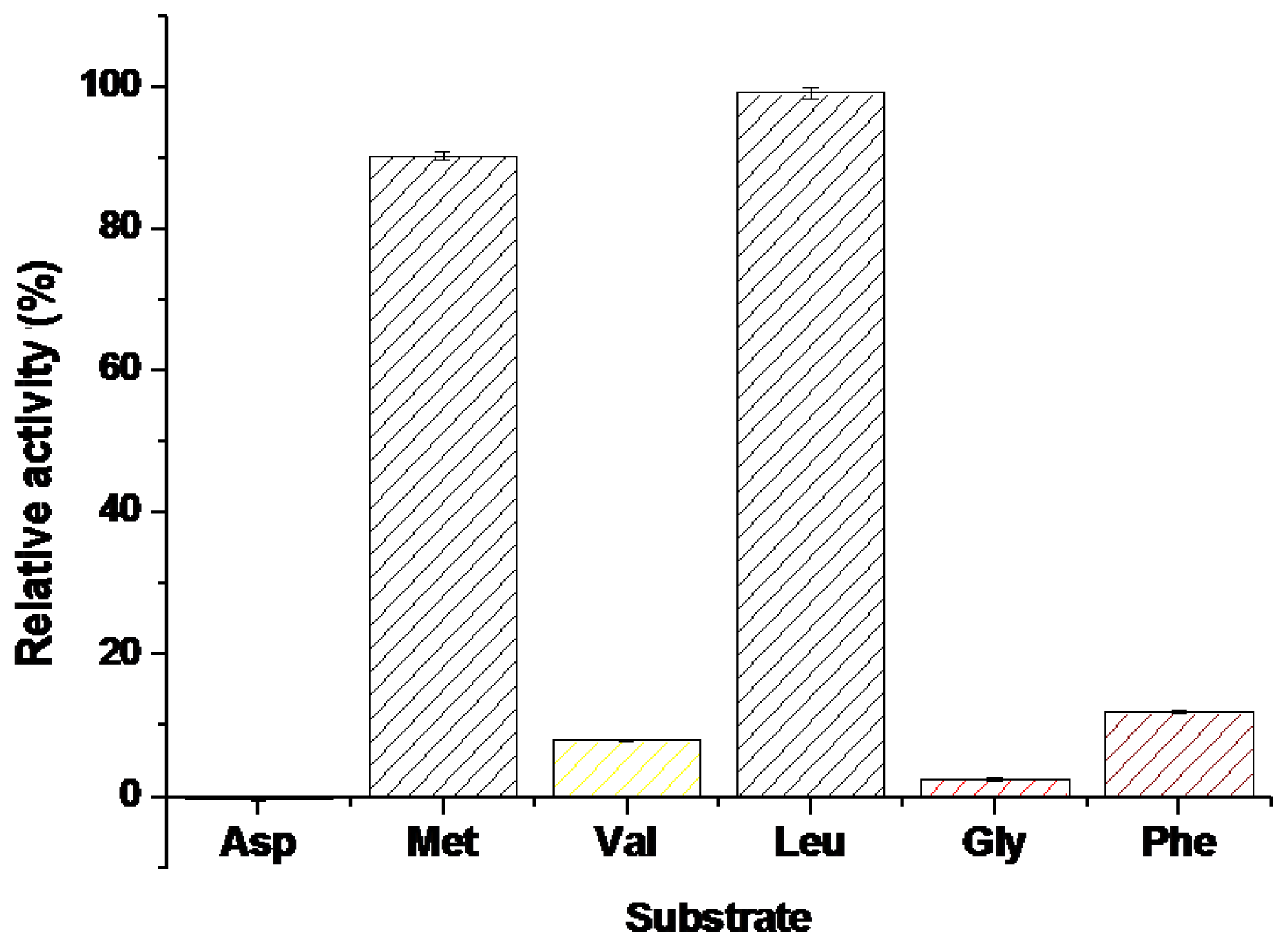
2.2.4. The Substrate Selectivity of BkLAP
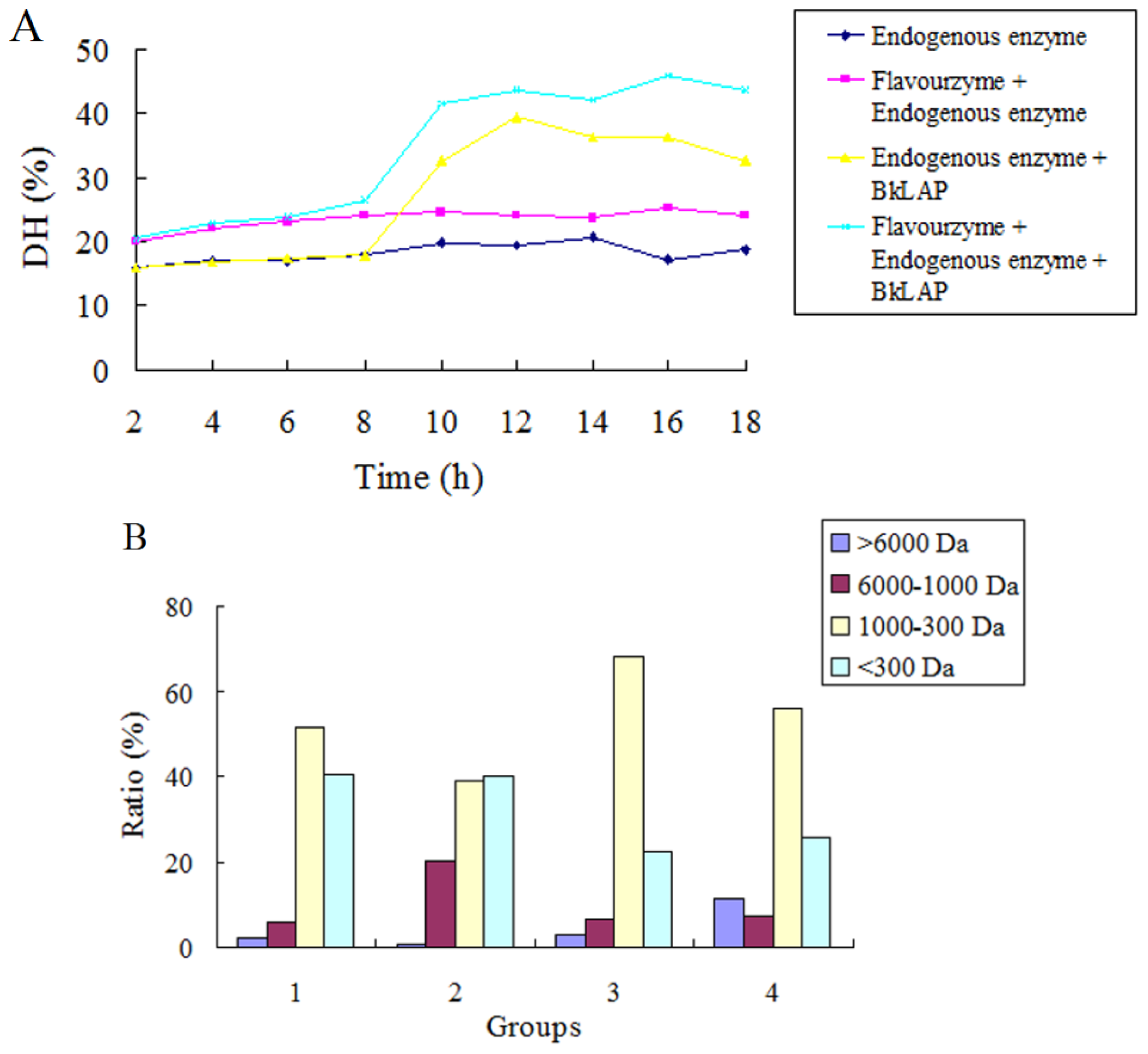
2.3. BkLAP Greatly Promoted the Hydrolysis of Anchovy Protein
3. Experimental Section
3.1. Total Gene Synthesis of Leucine Aminopeptidase and Construction of Expression Plasmid
3.2. Gene Expression and Enzyme Purification
3.3. Electrophoresis and Protein Assay
3.4. Enzyme Assay
3.5. Circular Dichroism Spectroscopy
3.6. Anchovy Hydrolytic Experiment
3.6.1. Materials
3.6.2. Production of Protein Hydrolysate with Commercial Enzymes Combined with BkLAP
3.6.3. Measurement of Degree of Hydrolysis (DH)
3.6.4. Free Amino Acid Analysis
3.6.5. Molecular Weight Distribution and Free Amino Acids Contents Anlaysis
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
References
- Eun, J.S.; Beauchemin, K.A.; Hong, S.H.; Bauer, M.W. Exogenous enzymes added to untreated or ammoniated rice straw: Effects on in vitro fermentation characteristics and degradability. Anim. Feed Sci. Technol 2006, 131, 87–102. [Google Scholar]
- EIBoushy, A.R.; van der Poel, A.F.B.; Walraven, O.E.D. Feather meal—A biological waste: Its processing and utilization as a feedstuff for poultry. Biol. Wastes 1990, 32, 39–74. [Google Scholar]
- Nodar, R.; Acea, M.J.; Carballas, T. Microbial compostion of poultry excreta. Biol. Wastes 1990, 33, 95–105. [Google Scholar]
- Urlings, H.A.; van Logtestijn, J.G.; Bijker, P.G. Slaughter by-products: Problems, preliminary research and possible solutions. Vet. Quart 1992, 14, 34–38. [Google Scholar]
- Urlings, H.A.; Fransen, N.G.; Bijker, P.G.; van Logtestijn, J.G. Proteolysis and amino acid breakdown of heated and irradiated poultry byproducts and muscle tissue. J. Anim. Sci 1993, 71, 2432–2438. [Google Scholar]
- Yu, J.; Ahmedna, M.; Goktepe, I. Peanut protein concentrate: Production and functional properties as affected by processing. Food Chem 2007, 103, 121–129. [Google Scholar]
- Guerard, F.; Dufosse, L.; Broise, D.D.L.; Binet, A. Enzymatic hydrolysis of proteins from yellowfin tuna (Thunnus albacares) wastes using Alcalase. J. Mol. Catal. B Enzym 2001, 11, 1051–1059. [Google Scholar]
- Quist, E.E.; Phillips, R.D.; Saalia, F.K. The effect of enzyme systems and processing on the hydrolysis of peanut (Arachis hypogaea L.) protein. LWT Food Sci. Technol 2009, 42, 1717–1721. [Google Scholar]
- Saha, B.C.; Hayashi, K. Debittering of protein hydrolyzates. Biotechnol. Adv 2001, 19, 355–370. [Google Scholar]
- Kim, H.; Lipscomb, W.N. Structure and mechanism of bovine lens leucine aminopeptidase. Adv. Enzymol. Areas Mol. Biol 1994, 68, 153–213. [Google Scholar]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial protease. Microbiol. Mol. Biol. Rev 1998, 62, 597–635. [Google Scholar]
- Kamphuis, J.; Meijer, E.M.; Boesten, W.H.J.; Broxterman, Q.B.; Kaptein, B.; Hermes, H.F.M.; Schoemaker, H.E. Production of natural and synthetic l- and d-amino acids by aminopeptidases and amino amidases. In Biocatalytic Production of Amino Acids and Derivatives; Rozzell, J.D., Wagner, F., Eds.; Wiley: New York, NY, USA, 1992; pp. 178–206. [Google Scholar]
- Toldra, F.; Aristoy, A.C.; Flores, M. Contribution of muscle aminopeptidase to flavor development in dry cured ham. Food Res. Int 2000, 33, 181–185. [Google Scholar]
- Lin, L.L.; Hsu, W.H.; Wu, C.P.; Chi, M.C.; Chou, W.M.; Hu, H.Y. A thermostable leucine aminopeptidase from Bacillus kaustophilus CCRC 11223. Extremophiles 2004, 8, 79–87. [Google Scholar]
- Chi, M.C.; Huang, H.B.; Liu, J.S.; Wang, W.C.; Liang, W.C.; Lin, L.L. Residues threonine346 and leucine 352 are critical for the proper function of Bacillus kaustophilus leucine aminopeptidase. FEMS Microbiol. Lett 2006, 260, 156–161. [Google Scholar]
- Chi, M.C.; Liu, J.S.; Wang, W.C.; Lin, L.L.; Huang, H.B. Site-directed mutagenesis of the conserved Ala348 and Gly350 residues at the putative active site of Bacillus kaustophilus aminopeptidase. Biochimie 2008, 90, 811–819. [Google Scholar]
- Magboul, A.A.A.; McSweeney, P.L.H. Purification and characterization of an aminopeptidase from Lactobacillus curvatus DPC2024. Int. Dairy J 1999, 9, 107–116. [Google Scholar]
- Montel, M.C.; Seronie, M.P.; Talon, R.; Hébraud, M. Purification and characterization of a dipeptidase from Lactobacillus sake. Appl. Environ. Microbiol 1995, 61, 837–839. [Google Scholar]
- Ramachandrin, L.K.; Witkop, B. The interaction of mercuric acetate with indoles, tryptophan and proteins. Biochemistry 1964, 3, 1603–1611. [Google Scholar]
- Xia, S.Y.; Deng, S.G.; Xie, C.; Huo, J.C. Nutritional ingredient and antihyperlipidemia action of protein hyrolysates from anchovy Engraulis japonicus. Oceanologia Et Limnologia Sini 2008, 39, 368–373. [Google Scholar]
- Siringan, P.; Raksakulthai, I.; Yongsawatdigul, J. Partial purification and characterization of trypsin-like proteinases in Indian anchovy. Food Chem 2007, 101, 82–89. [Google Scholar]
- Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Utilization of chickpea protein isolates for production of peptides with angiotensin I-converting enzyme (ACE)-inhibitory activity. J. Sci. Food Agric 2002, 82, 960–965. [Google Scholar]
- Plumb, G.W.; Mills, E.N.C.; Tatton, M.J.; Dursel, C.C.M.; Lambert, N.; Morgan, M.R.A. Effect of thermal and proteolytic processing on glycinin, the 11S globulin of soy (Glycine max): A study utilizing monoclonal and polyclonal antibodies. J. Agric. Food Chem 1994, 42, 834–840. [Google Scholar]
- Chiang, W.D.; Shih, C.J.; Chu, Y.H. Functional properties of soy protein hydrolysate produced from a continuous membrane reactor system. Food Chem 1999, 65, 189–194. [Google Scholar]
- Xiong, A.S.; Yao, Q.H.; Peng, R.H.; Li, X.; Fan, H.Q.; Cheng, Z.M.; Li, Y. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucl. Acids Res 2004, 32. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970, 227, 680–685. [Google Scholar]
- Kuo, L.Y.; Hwang, G.Y.; Lai, Y.J.; Yang, S.L.; Lin, L.L. Overexpression, purification, and characterization of the recombinant leucine aminopeptidase II of Bacillus stearothermophilus. Curr. Microbiol 2003, 47, 40–45. [Google Scholar]
- Nilsang, S.; Lertsiri, S.; Suphantharika, M.; Assavanig, A. Optimization of enzymatic hydrolysis of fish soluble concentrate by commercial proteases. J. Food Eng. 2005, 70, 571–578. [Google Scholar]
- Liu, C.; Wang, H.L.; Cui, Z.M.; He, X.L.; Wang, X.S.; Zeng, X.X.; Ma, H. Optimization of extraction and isolation for 11S and 7S globulins of soybean seed storage protein. Food Chem 2007, 102, 1310–1316. [Google Scholar]

| pH | Citrate-phosphate buffer | McIlvanie buffer | HEPES buffer | Potassium-phosphate buffer | Tris-HCl buffer | Glycine-NaOH buffer |
|---|---|---|---|---|---|---|
| 3.0 | 0.00 ± 0.00 | 0.06 ± 0.00 | — | — | — | — |
| 4.0 | 0.00 ± 0.00 | 0.10 ± 0.02 | — | — | — | — |
| 5.0 | 0.14 ± 0.07 | 0.49 ± 0.18 | — | — | — | — |
| 6.0 | 0.66 ± 0.08 | 0.75 ± 0.08 | — | 4.43 ± 0.56 | — | — |
| 7.0 | 0.31 ± 0.06 | 2.81 ± 2.79 | 44.70 ± 3.32 | 63.59 ± 3.05 | — | — |
| 7.5 | — | 12.38 ± 1.29 | 53.10 ± 2.15 | — | — | — |
| 8.0 | — | 56.26 ± 7.42 | 31.44 ± 6.67 | 100.00 ± 0.58 | 40.45 ± 0.94 | — |
| 9.0 | — | — | — | — | 39.32 ± 0.99 | 1.79 ± 1.63 |
| 10.0 | — | — | — | — | — | 1.86 ± 1.61 |
| 11.0 | — | — | — | — | — | 2.19 ± 1.65 |
| 12.0 | — | — | — | — | — | 0.00 ± 0.00 |
| pH 4.0 | pH 6.0 | pH 8.0 | pH 10.0 | |
|---|---|---|---|---|
| Helix | 43.87% a | 32.40% b | 27.67% c | 25.57% c |
| Sheet | 0.00% a | 37.27% b | 38.10% b | 40.27% b |
| Turn | 32.83% a | 1.53% b | 5.50% b | 7.30% b |
| Random | 23.27% a | 28.77% b | 28.67% b | 26.83% b |
| Metal ions | Concentration (M) | Relative activity (%) |
|---|---|---|
| None | — | 100 |
| Hg2+ | 1×10−4 | 6 |
| Mg2+ | 1×10−4 | 39 |
| Fe2+ | 1×10−4 | 99 |
| Fe3+ | 1×10−4 | 48 |
| Ca2+ | 1×10−4 | 32 |
| Ba2+ | 1×10−4 | 101 |
| Cu2+ | 1×10−4 | 48 |
| Zn2+ | 1×10−4 | 22 |
| Li2+ | 1×10−4 | 97 |
| Co2+ | 1×10−5 | 141 |
| 5×10−5 | 175 | |
| 1×10−4 | 202 | |
| 5×10−4 | 243 | |
| Mn2+ | 1×10−5 | 100 |
| 5×10−5 | 420 | |
| 1×10−4 | 491 | |
| 5×10−4 | 755 | |
| Ni2+ | 1×10−6 | 380 |
| 1×10−5 | 880 | |
| 5×10−5 | 1,007 | |
| 1×10−4 | 1,722 | |
| 5×10−4 | 3,500 |
| None | Co2+ | Ni2+ | Zn2+ | Cu2+ | Mn2+ | Hg2+ | Fe3+ | Ca2+ | |
|---|---|---|---|---|---|---|---|---|---|
| Helix | 27.67% a | 29.40% a | 30.73% a | 28.93% a | 30.33% a | 29.87% a | 37.07% b | 31.97% a | 28.23% a |
| Sheet | 38.10% a | 35.83% a | 36.47% a | 36.10% a | 35.23% a | 36.83% a | 0.00% b | 32.50% a | 37.77% a |
| Turn | 5.50% a | 6.20% a | 3.77% a | 5.87% a | 6.10% a | 4.43% a | 28.77% b | 6.67% a | 6.47% a |
| Random | 28.67% a | 28.60% a | 29.07% a | 29.07% a | 28.37% a | 28.83% a | 34.17% b | 28.87% a | 27.53% a |
| Groups | Enzyme | Temperature (°C) | pH |
|---|---|---|---|
| 1 | Endogenous enzyme | 55.0 | 7.0 |
| 2 | Flavourzyme + Endogenous enzyme | 50.0 | 6.5 |
| 3 | Endogenous enzyme + BkLAP | 55.0 °C for 6 h and then 70.0 °C for 12 h | 7.0 for 6 h and then 8.0 |
| 4 | Flavourzyme + Endogenous enzyme + BkLAP | 50.0 °C for 6 h and then 70.0 °C for 12 h | 6.5 for 6 h and then 8.0 |
| Group 1 | Group 2 | Group 3 | Group 4 | |
|---|---|---|---|---|
| Asp | 139.03 | 179.62 | 249.74 | 305.93 |
| Glu | 181.22 | 203.05 | 387.26 | 461.61 |
| Ser | 135.09 | 225.65 | 159.20 | 294.60 |
| Gly | 60.06 | 90.83 | 120.17 | 203.97 |
| His | 115.55 | 235.00 | 149.35 | 253.23 |
| Arg | 347.31 | 398.17 | 434.78 | 541.84 |
| Thr | 95.54 | 182.94 | 168.62 | 241.27 |
| Ala | 173.87 | 230.44 | 283.05 | 379.32 |
| Pro | 27.48 | 45.99 | 50.40 | 75.08 |
| Tyr | 85.78 | 104.84 | 782.47 | 887.89 |
| Val | 138.53 | 211.30 | 221.81 | 324.37 |
| Met | 95.27 | 129.20 | 125.29 | 169.00 |
| Cys | 5.88 | 15.40 | 39.56 | 58.87 |
| Ile | 131.57 | 197.37 | 198.73 | 285.70 |
| Leu | 268.26 | 372.34 | 388.64 | 530.67 |
| Phe | 137.05 | 174.54 | 187.47 | 268.58 |
| Lys | 300.50 | 362.33 | 447.30 | 541.61 |
| Total | 2,437.99 | 3,359.03 | 4,393.82 | 5,823.54 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shen, Y.; Wang, F.; Lan, D.; Liu, Y.; Yang, B.; Wang, Y. Biochemical Properties and Potential Applications of Recombinant Leucine Aminopeptidase from Bacillus kaustophilus CCRC 11223. Int. J. Mol. Sci. 2011, 12, 7609-7625. https://doi.org/10.3390/ijms12117609
Shen Y, Wang F, Lan D, Liu Y, Yang B, Wang Y. Biochemical Properties and Potential Applications of Recombinant Leucine Aminopeptidase from Bacillus kaustophilus CCRC 11223. International Journal of Molecular Sciences. 2011; 12(11):7609-7625. https://doi.org/10.3390/ijms12117609
Chicago/Turabian StyleShen, Yanfei, Fanghua Wang, Dongming Lan, Yuanyuan Liu, Bo Yang, and Yonghua Wang. 2011. "Biochemical Properties and Potential Applications of Recombinant Leucine Aminopeptidase from Bacillus kaustophilus CCRC 11223" International Journal of Molecular Sciences 12, no. 11: 7609-7625. https://doi.org/10.3390/ijms12117609




