Practical Application of Toxicogenomics for Profiling Toxicant-Induced Biological Perturbations
Abstract
:1. Introduction
2. Advancement of Toxicogenomics
2.1. Toxicogenomic Biomarker Gene Sets
2.2. Public and Commercial Microarray Database
2.3. Open Source Software
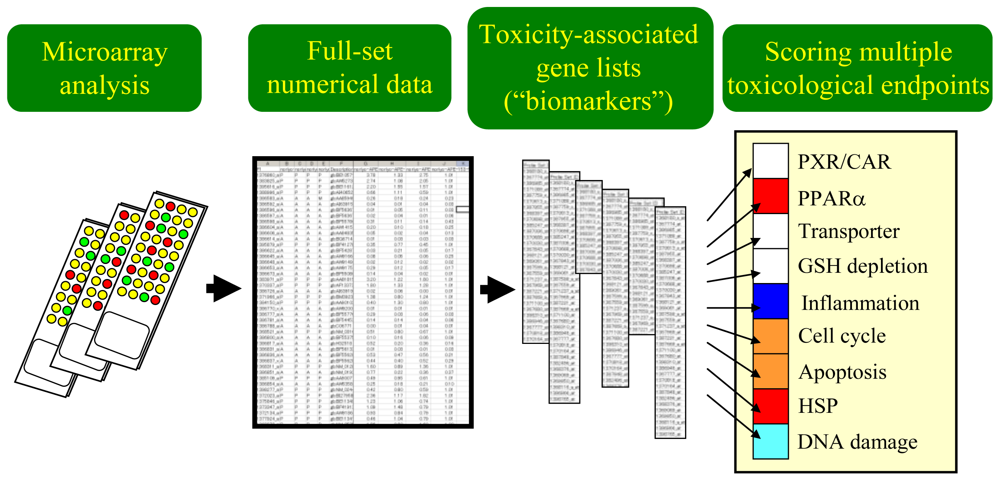
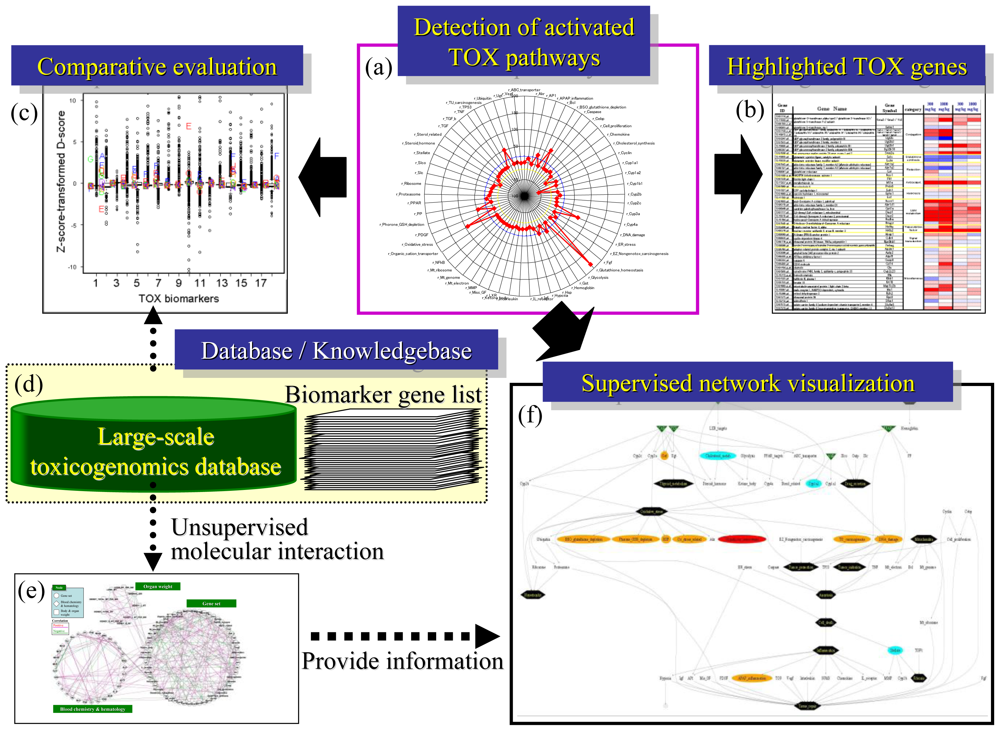
3. Practical Application of TGx Database and Biomarkers
3.1. Scoring the Gene Set-Level Expression Changes
3.2. Differentially-Regulated Gene Score (D-score)
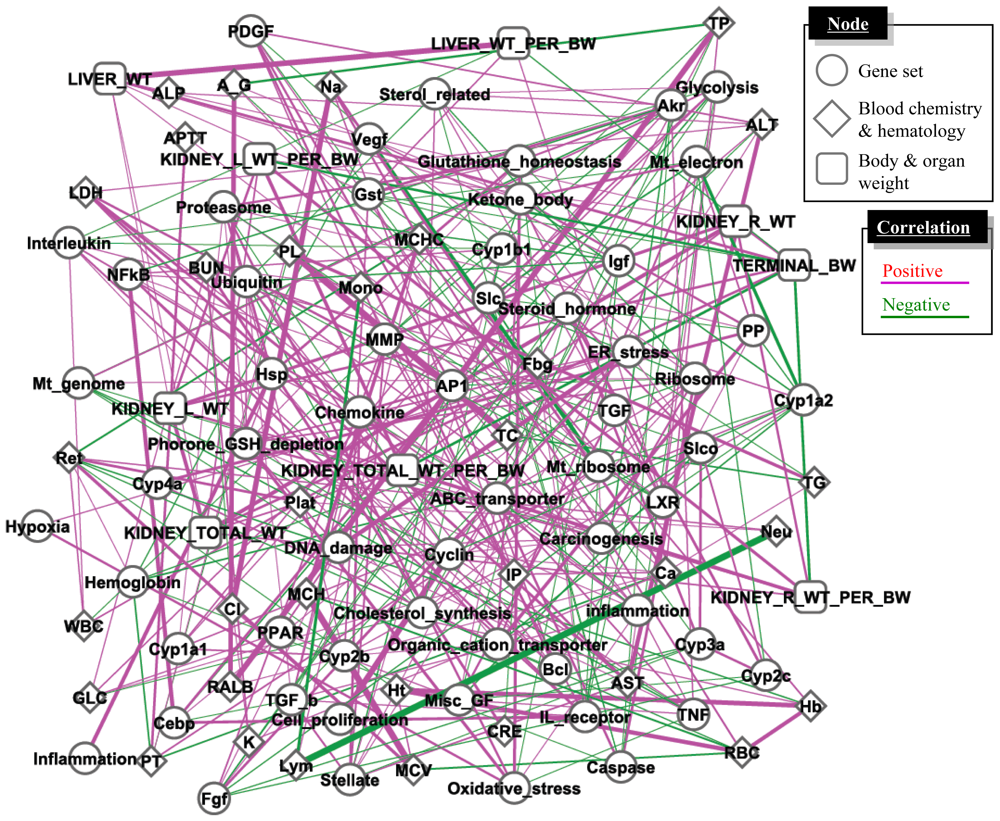
3.3. Inference of Gene Set-Level Network Structure Using a TGx Database
4. Case Study: Bromobenzene-Induced Molecular Perturbation
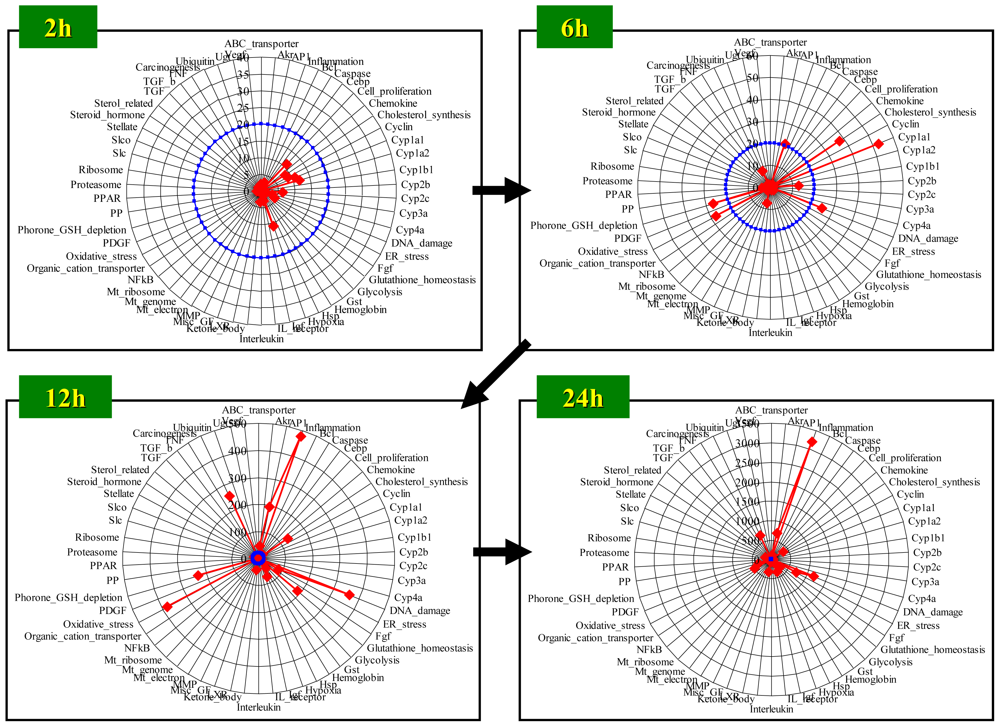
4.1. Radar Chart Presentation
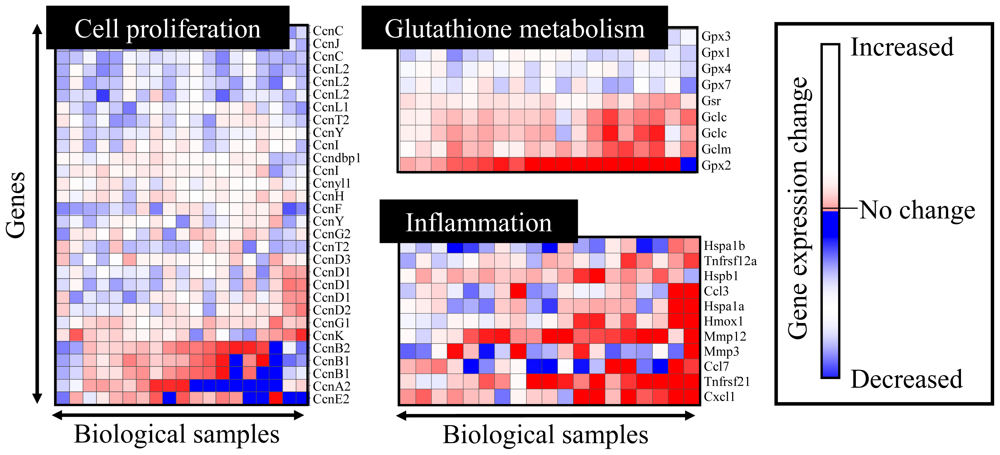
4.2. Heat Map Presentation
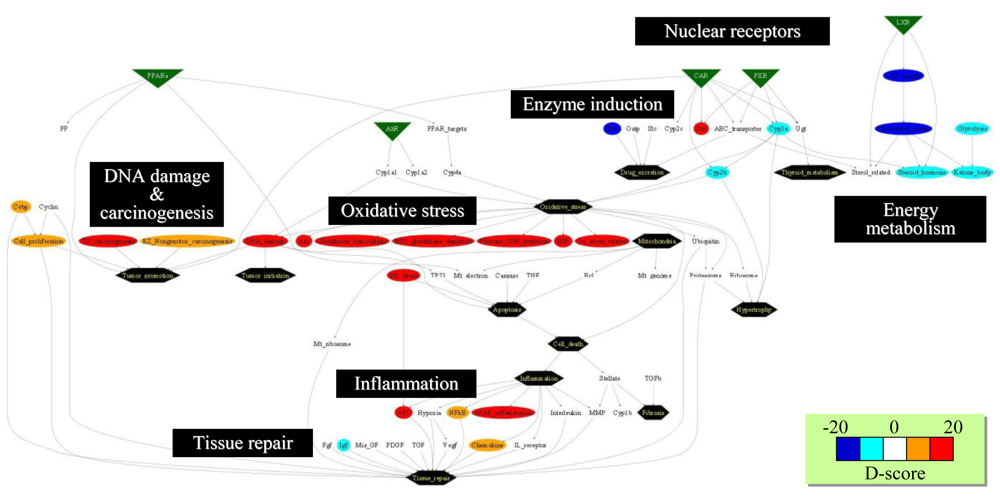
4.3. Supervised Network Structure Presentation
5. Conclusion
Acknowledgements
References
- Kaplowitz, N. Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov 2005, 4, 489–499. [Google Scholar]
- Marchetti, S; Mazzanti, R; Beijnen, JH; Schellens, JH. Concise review: Clinical relevance of drug drug and herb drug interactions mediated by the ABC transporter ABCB1 (MDR1, P-glycoprotein). Oncologist 2007, 12, 927–941. [Google Scholar]
- Haddad, A; Davis, M; Lagman, R. The pharmacological importance of cytochrome CYP3A4 in the palliation of symptoms: Review and recommendations for avoiding adverse drug interactions. Support. Care Cancer 2007, 15, 251–257. [Google Scholar]
- Kodama, S; Negishi, M. Phenobarbital confers its diverse effects by activating the orphan nuclear receptor car. Drug Metab. Rev 2006, 38, 75–87. [Google Scholar]
- Kitano, H. Towards a theory of biological robustness. Mol. Syst. Biol 2007, 3, 137. [Google Scholar]
- Boverhof, DR; Zacharewski, TR. Toxicogenomics in risk assessment: applications and needs. Toxicol. Sci 2006, 89, 352–360. [Google Scholar]
- Waring, JF; Jolly, RA; Ciurlionis, R; Lum, PY; Praestgaard, JT; Morfitt, DC; Buratto, B; Roberts, C; Schadt, E; Ulrich, RG. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol. Appl. Pharmacol 2001, 175, 28–42. [Google Scholar]
- Waring, JF; Ciurlionis, R; Jolly, RA; Heindel, M; Ulrich, RG. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol. Lett 2001, 120, 359–368. [Google Scholar]
- Ashburner, M; Ball, CA; Blake, JA; Botstein, D; Butler, H; Cherry, JM; Davis, AP; Dolinski, K; Dwight, SS; Eppig, JT; Harris, MA; Hill, DP; Issel-Tarver, L; Kasarskis, A; Lewis, S; Matese, JC; Richardson, JE; Ringwald, M; Rubin, GM; Sherlock, G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 2000, 25, 25–29. [Google Scholar]
- Goto, S; Bono, H; Ogata, H; Fujibuchi, W; Nishioka, T; Sato, K; Kanehisa, M. Organizing and computing metabolic pathway data in terms of binary relations. Pac. Symp. Biocomput 1997, 175–186. [Google Scholar]
- Dahlquist, KD; Salomonis, N; Vranizan, K; Lawlor, SC; Conklin, BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet 2002, 31, 19–20. [Google Scholar]
- Kiyosawa, N; Ando, Y; Manabe, S; Yamoto, T. Toxicogenomic biomarkers for liver toxicity. J. Toxicol. Pathol 2009, 22, 35–52. [Google Scholar]
- Kier, LD; Neft, R; Tang, L; Suizu, R; Cook, T; Onsurez, K; Tiegler, K; Sakai, Y; Ortiz, M; Nolan, T; Sankar, U; Li, AP. Applications of microarrays with toxicologically relevant genes (tox genes) for the evaluation of chemical toxicants in Sprague Dawley rats in vivo and human hepatocytes in vitro. Mutat. Res 2004, 549, 101–113. [Google Scholar]
- Ellinger-Ziegelbauer, H; Gmuender, H; Bandenburg, A; Ahr, HJ. Prediction of a carcinogenic potential of rat hepatocarcinogens using toxicogenomics analysis of short-term in vivo studies. Mutat. Res 2008, 637, 23–39. [Google Scholar]
- Uehara, T; Hirode, M; Ono, A; Kiyosawa, N; Omura, K; Shimizu, T; Mizukawa, Y; Miyagishima, T; Nagao, T; Urushidani, T. A toxicogenomics approach for early assessment of potential non-genotoxic hepatocarcinogenicity of chemicals in rats. Toxicology 2008, 250, 15–26. [Google Scholar]
- Sawada, H; Takami, K; Asahi, SA. Toxicogenomic approach to drug-induced phospholipidosis: Analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol. Sci 2005, 83, 282–292. [Google Scholar]
- Hirode, M; Ono, A; Miyagishima, T; Nagao, T; Ohno, Y; Urushidani, T. Gene expression profiling in rat liver treated with compounds inducing phospholipidosis. Toxicol. Appl. Pharmacol 2008, 229, 290–299. [Google Scholar]
- Kiyosawa, N; Ito, K; Sakuma, K; Niino, N; Kanbori, M; Yamoto, T; Manabe, S; Matsunuma, N. Evaluation of glutathione deficiency in rat livers by microarray analysis. Biochem. Pharmacol 2004, 68, 1465–1475. [Google Scholar]
- Kiyosawa, N; Uehara, T; Gao, W; Omura, K; Hirode, M; Shimizu, T; Mizukawa, Y; Ono, A; Miyagishima, T; Nagao, T; Urushidani, T. Identification of glutathione depletion-responsive genes using phorone-treated rat liver. J. Toxicol. Sci 2007, 32, 469–486. [Google Scholar]
- Edgar, R; Domrachev, M; Lash, AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic. Acids Res 2002, 30, 207–210. [Google Scholar]
- Brazma, A; Parkinson, H; Sarkans, U; Shojatalab, M; Vilo, J; Abeygunawardena, N; Holloway, E; Kapushesky, M; Kemmeren, P; Lara, GG; Oezcimen, A; Rocca-Serra, P; Sansone, SA. Array Express—A public repository for microarray gene expression data at the EBI. Nucleic Acids Res 2003, 31, 68–71. [Google Scholar]
- Waters, M; Stasiewicz, S; Merrick, BA; Tomer, K; Bushel, P; Paules, R; Stegman, N; Nehls, G; Yost, KJ; Johnson, CH; Gustafson, SF; Xirasagar, S; Xiao, N; Huang, CC; Boyer, P; Chan, DD; Pan, Q; Gong, H; Taylor, J; Choi, D; Rashid, A; Ahmed, A; Howle, R; Selkirk, J; Tennant, R; Fostel, J. CEBS—Chemical Effects in Biological Systems: A public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic Acids Res 2008, 36, D892–D900. [Google Scholar]
- Mattingly, CJ; Rosenstein, MC; Davis, AP; Colby, GT; Forrest, JN, Jr; Boyer, JL. The comparative toxicogenomics database: A cross-species resource for building chemical-gene interaction networks. Toxicol. Sci 2006, 92, 587–595. [Google Scholar]
- Hayes, KR; Vollrath, AL; Zastrow, GM; McMillan, BJ; Craven, M; Jovanovich, S; Rank, DR; Penn, S; Walisser, JA; Reddy, JK; Thomas, RS; Bradfield, CA. EDGE: A centralized resource for the comparison, analysis, and distribution of toxicogenomic information. Mol. Pharmacol 2005, 67, 1360–1368. [Google Scholar]
- Gehlenborg, N; O’Donoghue, SI; Baliga, NS; Goesmann, A; Hibbs, MA; Kitano, H; Kohlbacher, O; Neuweger, H; Schneider, R; Tenenbaum, D; Gavin, AC. Visualization of omics data for systems biology. Nat. Methods 2010, 7, S56–S68. [Google Scholar]
- Gentleman, RC; Carey, VJ; Bates, DM; Bolstad, B; Dettling, M; Dudoit, S; Ellis, B; Gautier, L; Ge, Y; Gentry, J; Hornik, K; Hothorn, T; Huber, W; Iacus, S; Irizarry, R; Leisch, F; Li, C; Maechler, M; Rossini, AJ; Sawitzki, G; Smith, C; Smyth, G; Tierney, L; Yang, JY; Zhang, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 2004, 5, R80. [Google Scholar]
- R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Australia, 2008.
- Shannon, P; Markiel, A; Ozier, O; Baliga, NS; Wang, JT; Ramage, D; Amin, N; Schwikowski, B; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13, 2498–2504. [Google Scholar]
- Urushidani, T. Sahu, S, Ed.; Prediction of hepatotoxicity based on the toxicogenomics database. In Hepatotoxicity from Genomics to in Vitro and in Vivo Models; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 507–529. [Google Scholar]
- Kiyosawa, N; Shiwaku, K; Hirode, M; Omura, K; Uehara, T; Shimizu, T; Mizukawa, Y; Miyagishima, T; Ono, A; Nagao, T; Urushidani, T. Utilization of a one-dimensional score for surveying chemical-induced changes in expression levels of multiple biomarker gene sets using a large-scale toxicogenomics database. J. Toxicol. Sci 2006, 31, 433–448. [Google Scholar]
- Kiyosawa, N; Ando, Y; Watanabe, K; Niino, N; Manabe, S; Yamoto, T. Scoring multiple toxicological endpoints using a toxicogenomic database. Toxicol. Lett 2009, 188, 91–97. [Google Scholar]
- Mah, N; Thelin, A; Lu, T; Nikolaus, S; Kuhbacher, T; Gurbuz, Y; Eickhoff, H; Kloppel, G; Lehrach, H; Mellgard, B; Costello, CM; Schreiber, S. A comparison of oligonucleotide and cDNA-based microarray systems. Physiol. Genomics 2004, 16, 361–370. [Google Scholar]
- Severgnini, M; Bicciato, S; Mangano, E; Scarlatti, F; Mezzelani, A; Mattioli, M; Ghidoni, R; Peano, C; Bonnal, R; Viti, F; Milanesi, L; De Bellis, G; Battaglia, C. Strategies for comparing gene expression profiles from different microarray platforms: application to a case-control experiment. Anal. Biochem 2006, 353, 43–56. [Google Scholar]
- Waring, JF; Ulrich, RG; Flint, N; Morfitt, D; Kalkuhl, A; Staedtler, F; Lawton, M; Beekman, JM; Suter, L. Interlaboratory evaluation of rat hepatic gene expression changes induced by methapyrilene. Environ. Health Perspect 2004, 112, 439–448. [Google Scholar]
- Chu, TM; Deng, S; Wolfinger, R; Paules, RS; Hamadeh, HK. Cross-site comparison of gene expression data reveals high similarity. Environ. Health Perspect 2004, 112, 449–455. [Google Scholar]
- Fielden, MR; Nie, A; McMillian, M; Elangbam, CS; Trela, BA; Yang, Y; Dunn, RT, II; Dragan, Y; Fransson-Stehen, R; Bogdanffy, M; Adams, SP; Foster, WR; Chen, SJ; Rossi, P; Kasper, P; Jacobson-Kram, D; Tatsuoka, KS; Wier, PJ; Gollub, J; Halbert, DN; Roter, A; Young, JK; Sina, JF; Marlowe, J; Martus, HJ; Aubrecht, J; Olaharski, AJ; Roome, N; Nioi, P; Pardo, I; Snyder, R; Perry, R; Lord, P; Mattes, W; Car, BD. Interlaboratory evaluation of genomic signatures for predicting carcinogenicity in the rat. Toxicol. Sci 2008, 103, 28–34. [Google Scholar]
- Beyer, RP; Fry, RC; Lasarev, MR; McConnachie, LA; Meira, LB; Palmer, VS; Powell, CL; Ross, PK; Bammler, TK; Bradford, BU; Cranson, AB; Cunningham, ML; Fannin, RD; Higgins, GM; Hurban, P; Kayton, RJ; Kerr, KF; Kosyk, O; Lobenhofer, EK; Sieber, SO; Vliet, PA; Weis, BK; Wolfinger, R; Woods, CG; Freedman, JH; Linney, E; Kaufmann, WK; Kavanagh, TJ; Paules, RS; Rusyn, I; Samson, LD; Spencer, PS; Suk, W; Tennant, RJ; Zarbl, H. Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol. Sci 2007, 99, 326–337. [Google Scholar]
- Baker, VA; Harries, HM; Waring, JF; Duggan, CM; Ni, HA; Jolly, RA; Yoon, LW; de Souza, AT; Schmid, JE; Brown, RH; Ulrich, RG; Rockett, JC. Clofibrate-induced gene expression changes in rat liver: A cross-laboratory analysis using membrane cDNA arrays. Environ. Health Perspect 2004, 112, 428–438. [Google Scholar]
- Ulrich, RG; Rockett, JC; Gibson, GG; Pettit, SD. Overview of an interlaboratory collaboration on evaluating the effects of model hepatotoxicants on hepatic gene expression. Environ. Health Perspect 2004, 112, 423–427. [Google Scholar]
- Manoli, T; Gretz, N; Grone, HJ; Kenzelmann, M; Eils, R; Brors, B. Group testing for pathway analysis improves comparability of different microarray datasets. Bioinformatics 2006, 22, 2500–2506. [Google Scholar]
- Ma’ayan, A. Insights into the organization of biochemical regulatory networks using graph theory analyses. J. Biol. Chem 2009, 284, 5451–5455. [Google Scholar]
- Kiyosawa, N; Manabe, S; Sanbuissho, A; Yamoto, T. Gene set-level network analysis using a toxicogenomics database. Genomics 2010, 96, 39–49. [Google Scholar]
- Crzegorczyk, M; Husmeier, D; Werhli, AV. Emmert-Streib, F, Dehmer, M, Eds.; Reverse engineering gene regulatory networks with various machine learning methods. In Analysis of Microarray Data—A Network Approach; Wiley-VCH: Hoboken, NJ, USA, 2008. [Google Scholar]
- Lau, SS; Monks, TJ. The contribution of bromobenzene to our current understanding of chemically-induced toxicities. Life Sci 1988, 42, 1259–1269. [Google Scholar]


© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kiyosawa, N.; Manabe, S.; Yamoto, T.; Sanbuissho, A. Practical Application of Toxicogenomics for Profiling Toxicant-Induced Biological Perturbations. Int. J. Mol. Sci. 2010, 11, 3397-3412. https://doi.org/10.3390/ijms11093397
Kiyosawa N, Manabe S, Yamoto T, Sanbuissho A. Practical Application of Toxicogenomics for Profiling Toxicant-Induced Biological Perturbations. International Journal of Molecular Sciences. 2010; 11(9):3397-3412. https://doi.org/10.3390/ijms11093397
Chicago/Turabian StyleKiyosawa, Naoki, Sunao Manabe, Takashi Yamoto, and Atsushi Sanbuissho. 2010. "Practical Application of Toxicogenomics for Profiling Toxicant-Induced Biological Perturbations" International Journal of Molecular Sciences 11, no. 9: 3397-3412. https://doi.org/10.3390/ijms11093397



