Altered Expression of Signaling Genes in Jurkat Cells upon FTY720 Induced Apoptosis
Abstract
:1. Introduction
2. Results and Discussion
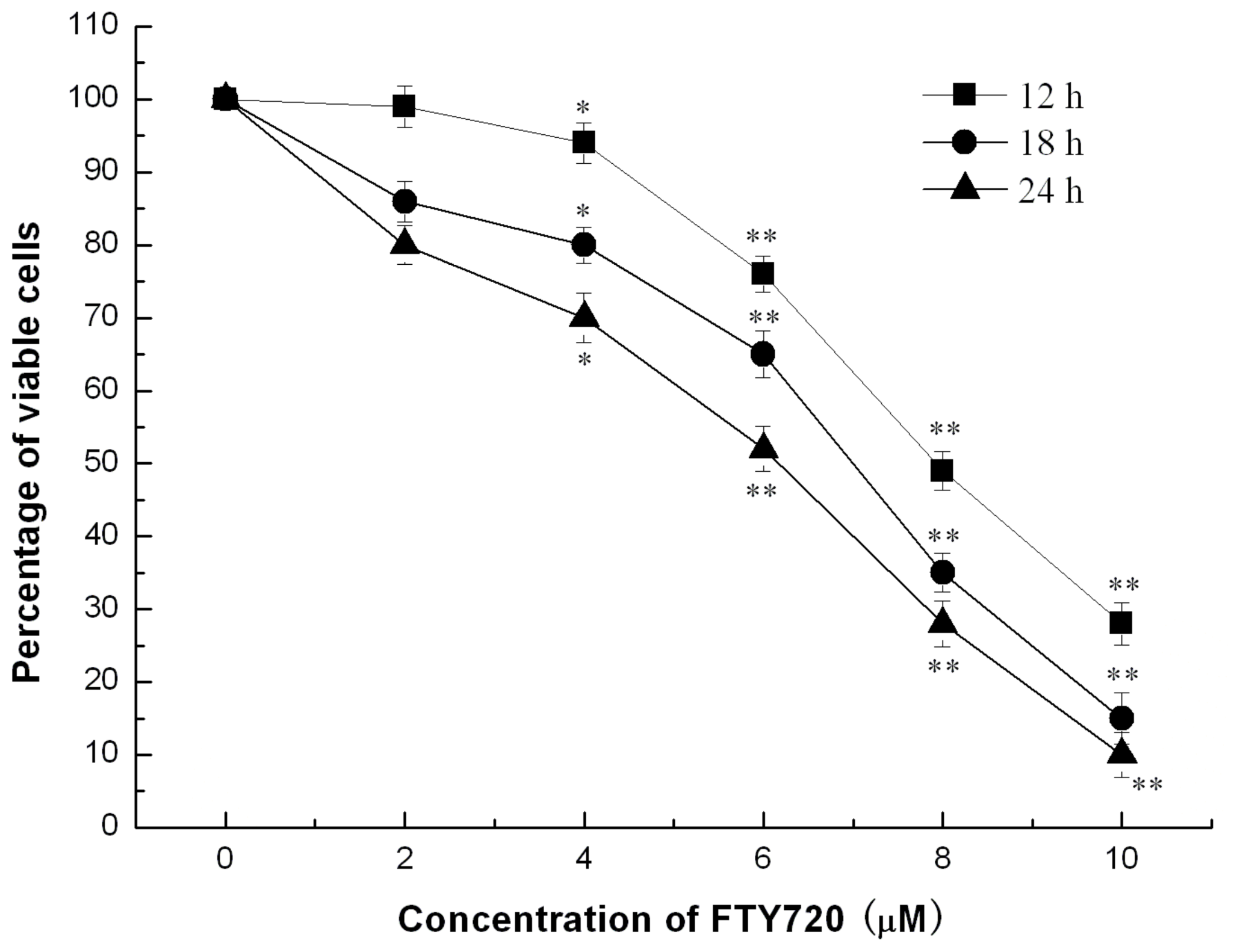
2.1. FTY720 Reduces Cell Viability
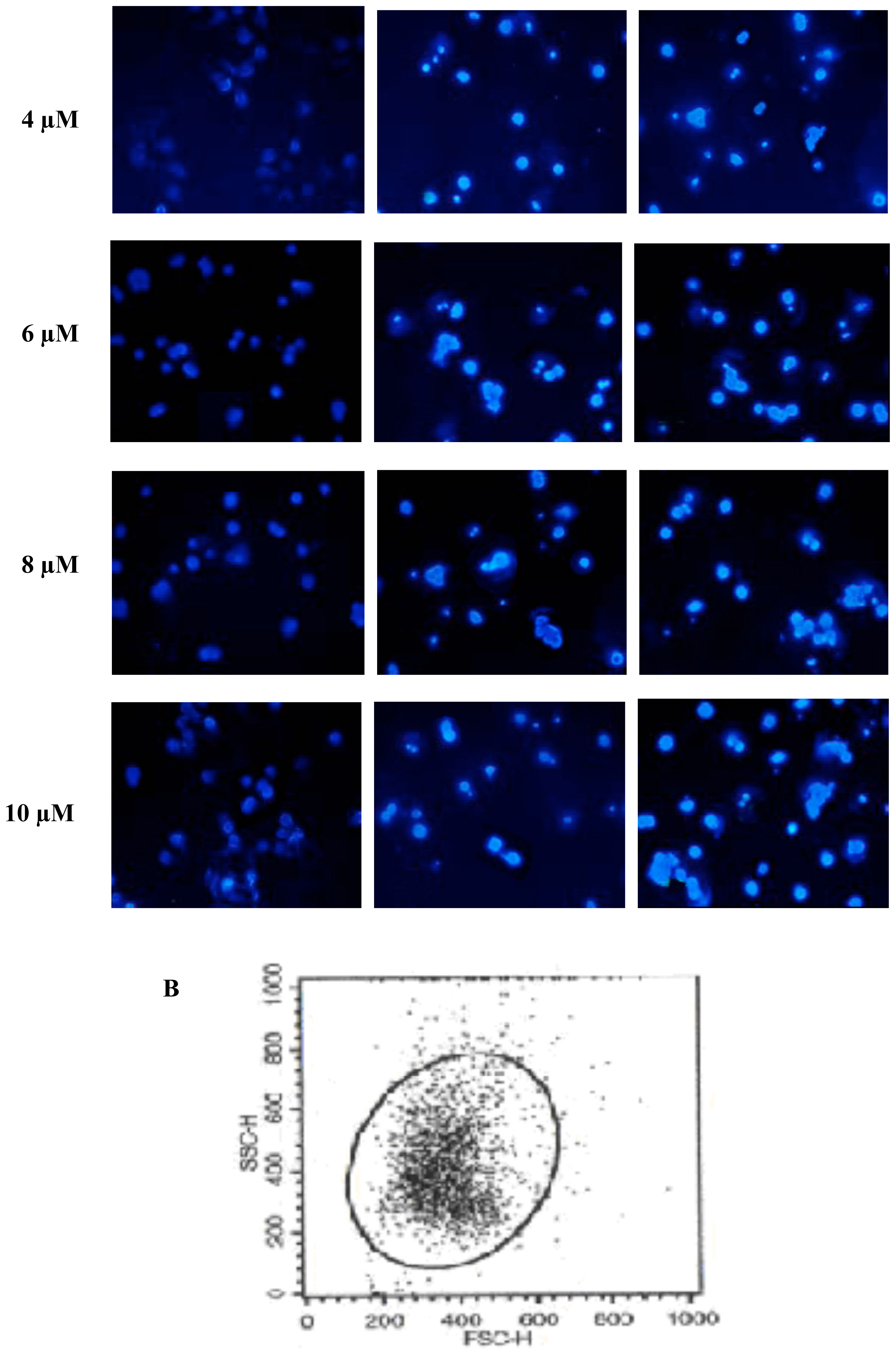
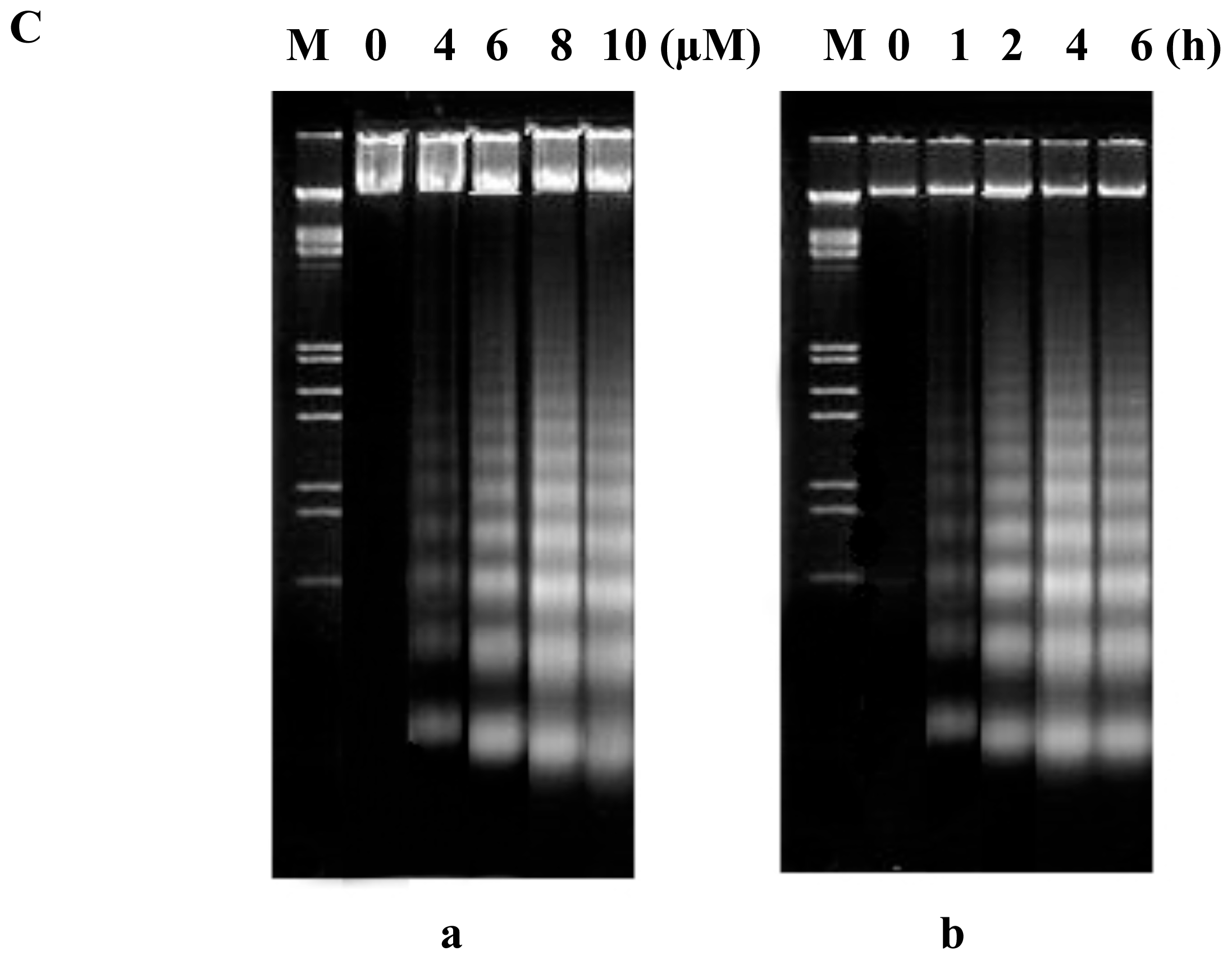
2.2. FTY720 Induces Cell Apoptosis in Jurkat Cells
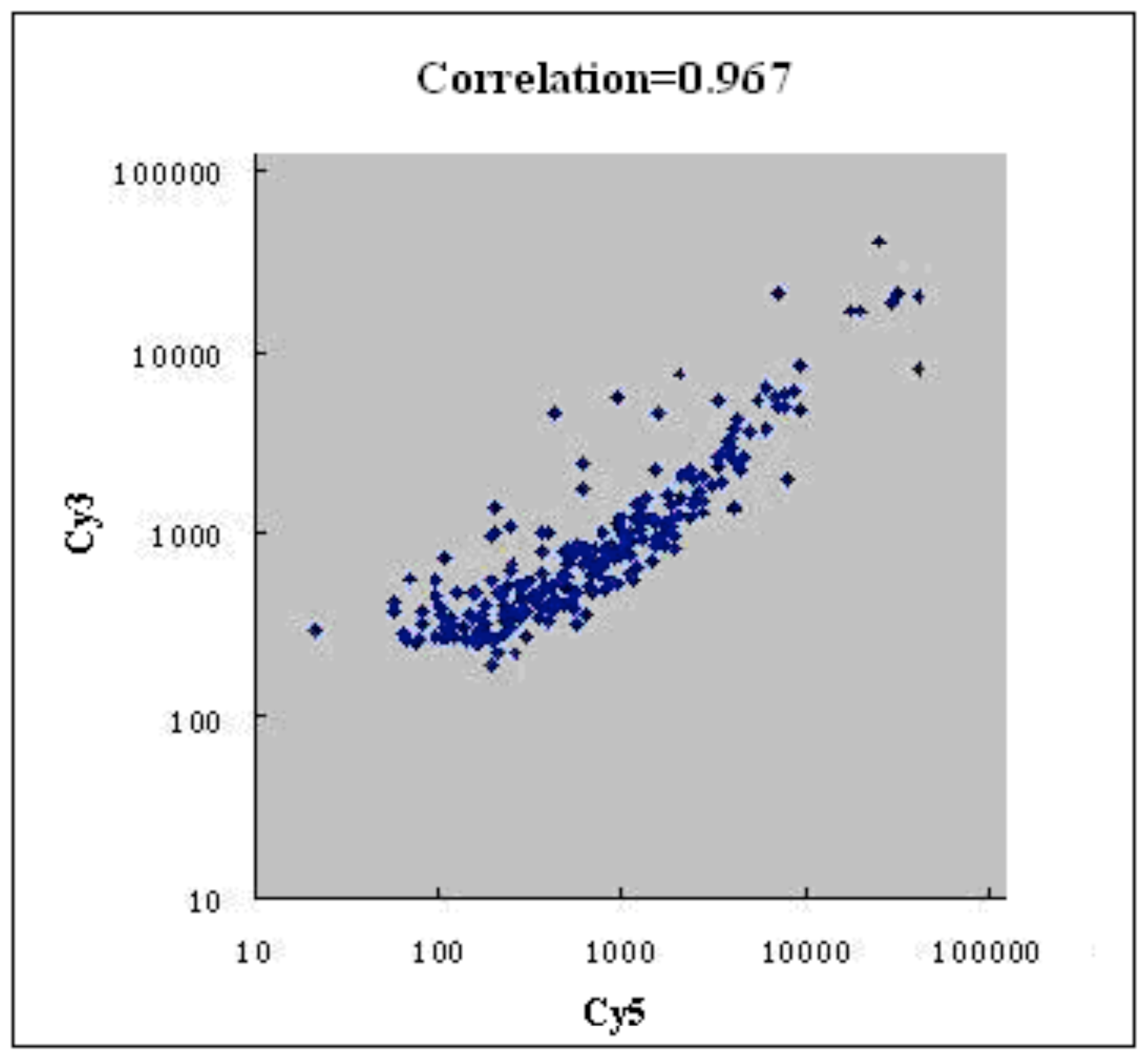
2.3. Microarray Analysis
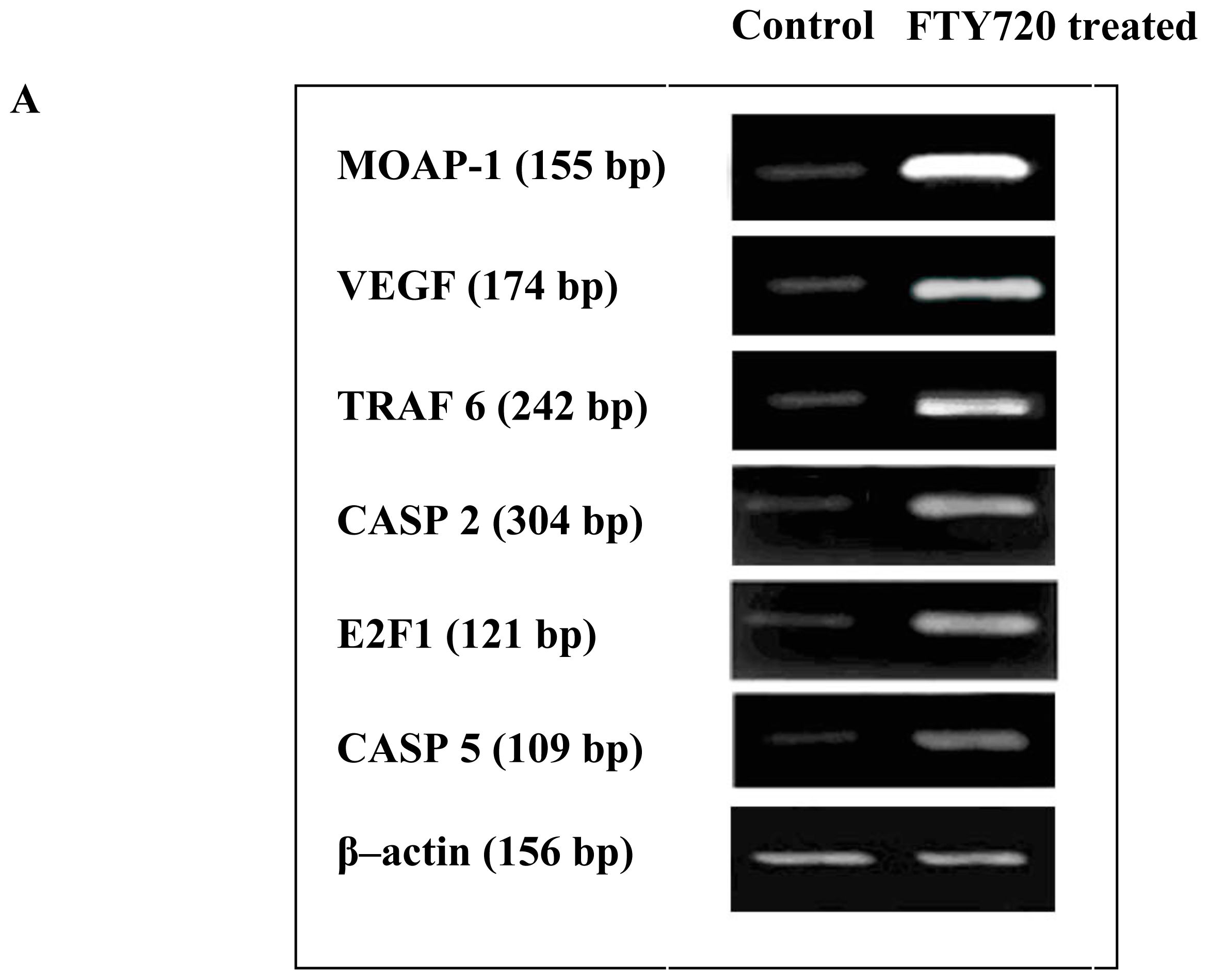
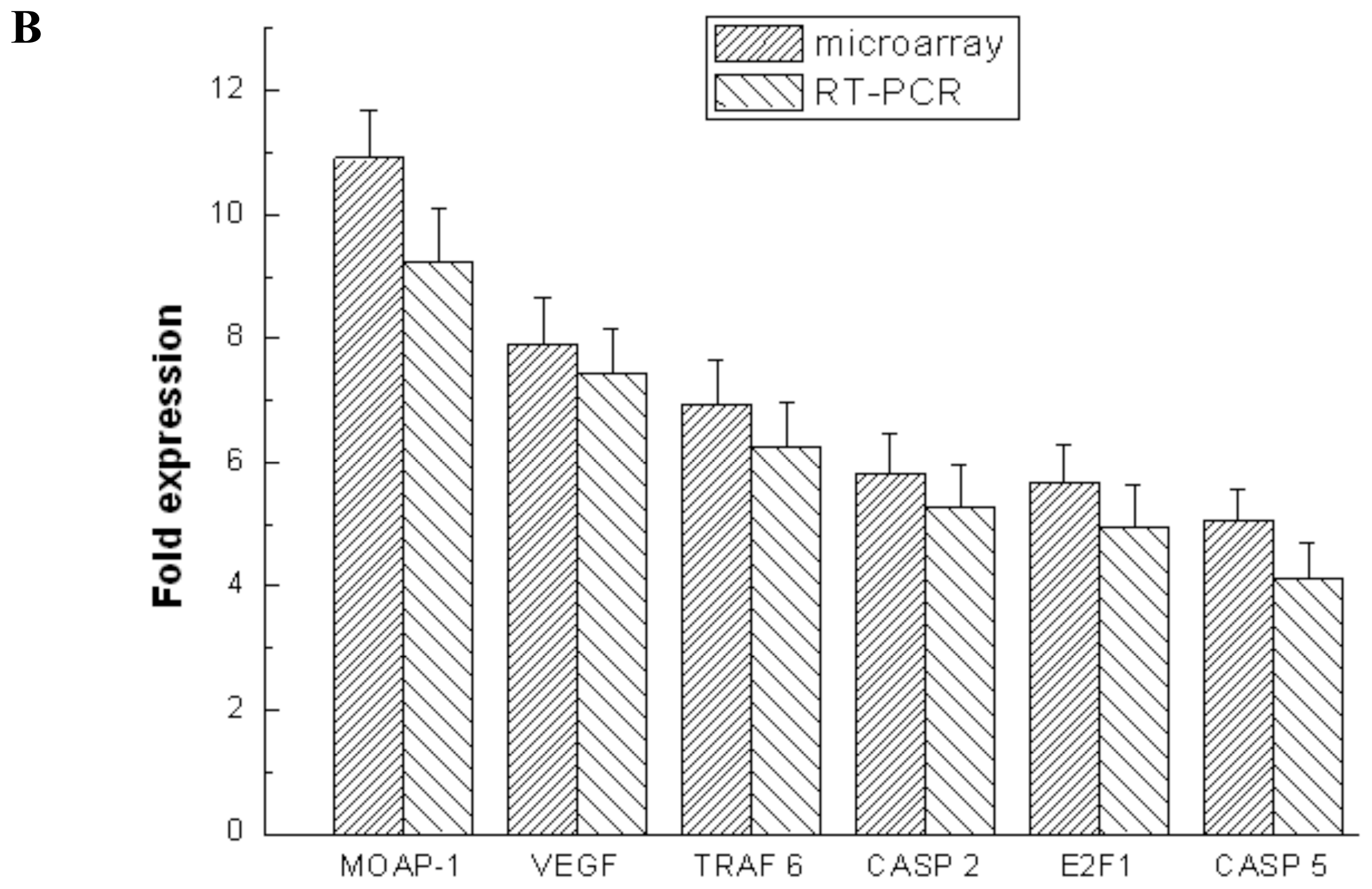
2.4. Validation of Microarray Data with RT-PCR and Western Blotting
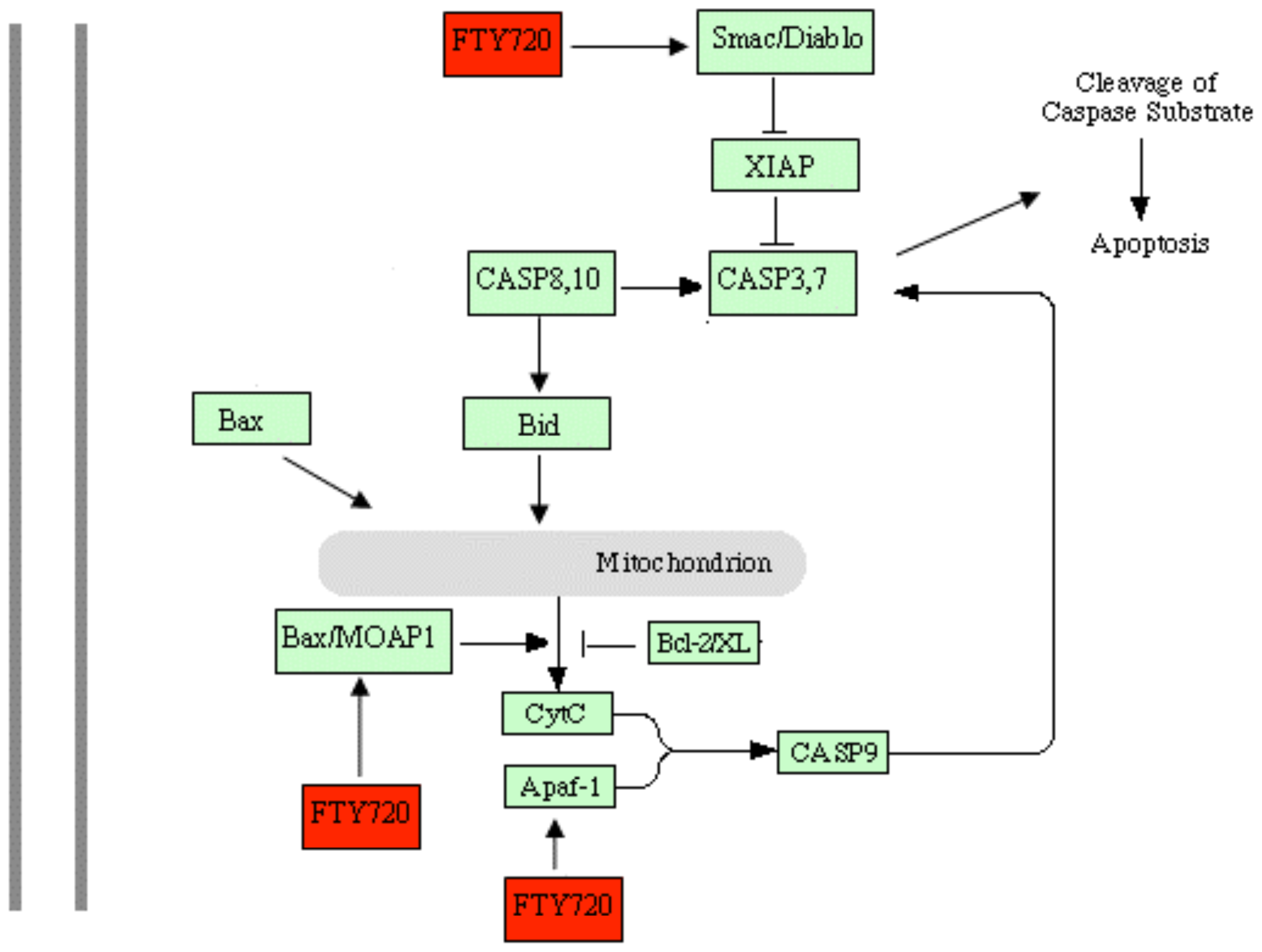
2.5. Discussion
3. Experimental Section
3.1. Materials
3.2. Cell Culture and FTY720 Treatment
3.3. Viability Assay by MTT
3.4. Apoptosis AssaysHoechst 33258 staining
Flow cytometry analysis
DNA fragmentation
3.5. Microarray Analysis
3.6. Semi-Quantitative RT-PCR Analysis
3.7. Western Blotting Analysis
3.8. Statistical Analysis
4. Conclusions
Acknowledgements
References and Notes
- Fujita, T; Inoue, K; Yamamoto, S; Ikumoto, T; Sasaki, S; Toyama, R; Chiba, K; Hoshino, Y; Okumoto, T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J. Antibiot 1994, 47, 208–215. [Google Scholar]
- Nagahara, Y; Enosawa, S; Ikekita, M; Suzuki, S; Shinomiya, T. Evidence that FTY720 induces T cell apoptosis in vivo. Immunopharmacology 2000, 48, 75–85. [Google Scholar]
- Gräler, MH; Goetzl, EJ. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G protein-coupled receptors. FASEB J 2004, 18, 551–553. [Google Scholar]
- Kataoka, H; Sugahara, K; Shimano, K; Teshima, K; Koyama, M; Fukunari, A; Chiba, K. FTY720, sphingosine 1-phosphate receptor modulator, ameliorates experimental autoimmune encephalomyelitis by inhibition of T cell infiltration. Cell Mol. Immunol 2005, 2, 439–448. [Google Scholar]
- Brinkmann, V. FTY720 mechanism of action and potential benefit in organ transplantation. Yonsei Med. J 2004, 45, 991–997. [Google Scholar]
- Chiba, K; Yanagawa, Y; Masubuchi, Y; Kataoka, H; Kawaguchi, T; Ohtsuki, M; Hoshino, Y. FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. J. Immunol 1998, 160, 5037–5044. [Google Scholar]
- Luo, ZJ; Tanaka, T; Kimura, F; Miyasaka, M. Analysis of the mode of action of a novel immunosuppressant FTY720 in mice. Immunopharmacology 1999, 41, 199–197. [Google Scholar]
- Henning, G; Ohl, L; Junt, T; Reiterer, P; Brinkmann, V; Nakano, H; Hohenberger, W; Lipp, M; Förster, R. CC chemokine receptor 7-dependent and –independent pathways for lymphocyte homing: Modulation by FTY720. J. Exp. Med 2001, 194, 1875–1881. [Google Scholar]
- Enosawa, S; Suzuki, S; Kakefuda, T; Li, XK; Amemiya, H. Induction of selective cell death targeting on mature T-lymphocytes in rats by a novel immunosuppressant, FTY720. Immunopharmacology 1996, 34, 171–179. [Google Scholar]
- Fujino, M; Funeshima, N; Kitazawa, Y; Kimura, H; Amemiya, H; Suzuki, S; Li, XK. Amelioration of experimental autoimmune encephalomyelitis in Lewis rats by FTY720 treatment. J. Pharmacol. Exp. Ther 2003, 305, 70–77. [Google Scholar]
- Azuma, H; Horie, S; Muto, S; Otsuki, Y; Matsumoto, K; Morimoto, J; Gotoh, R; Okuyama, A; Suzuki, S; Katsuoka, Y; Takahara, S. Selective cancer cell apoptosis induced by FTY720; evidence for a Bcl-dependent pathway and impairment in ERK activity. Anticancer Res 2003, 23, 3183–3193. [Google Scholar]
- Matsuda, T; Nakajima, H; Fujiwara, I; Mizuta, N; Oka, T. Caspase requirement for the apoptotic death of WR19L-induced by FTY720. Transplant Proc 1998, 30, 2355–2357. [Google Scholar]
- Matsuda, S; Minowa, A; Suzuki, S; Koyasu, S. Differential activation of c-Jun NH2-terminal kinase and p38 pathways during FTY720-induced apoptosis of T lymphocytes that is suppressed by the extracellular signal-regulated kinase pathway. J. Immunol 1999, 162, 3321–3326. [Google Scholar]
- Suzuki, S; Li, XK; Enosawa, S; Shinomiya, T. An immunosuppressive agent, FTY720, increases intracellular concentration of calcium ion and induces apoptosis in HL-60. Immunology 1997, 91, 594–600. [Google Scholar]
- Wang, JD; Takahara, S; Nonomura, N; Ichimaru, N; Toki, K; Azuma, H; Matsumiya, K; Okuyama, A; Suzuki, S. Early induction of apoptosis in androgen-independent prostate cancer cell line by FTY720 requires caspase-3 activation. Prostate 1999, 40, 50–55. [Google Scholar]
- Azuma, H; Takahara, S; Ichimaru, N; Wang, JD; Itoh, Y; Otsuki, Y; Morimoto, J; Fukui, R; Hoshiga, M; Ishihara, T; Nonomura, N; Suzuki, S; Okuyama, A; Katsuoka, Y. Marked prevention of tumor growth and metastasis by a novel immunosuppressive agent, FTY720, in mouse breast cancer models. Cancer Res 2002, 62, 1410–1419. [Google Scholar]
- Suzuki, S; Li, XK; Enosawa, S; Shinomiya, T. A new immunosuppressant, FTY720, induces bcl-2-associated apoptotic cell death in human lymphocytes. Immunology 1996, 89, 518–523. [Google Scholar]
- Suzuki, S; Li, XK; Shinomiya, T; Enosawa, S; Amemiya, H; Amari, M; Naoe, S. The in vivo induction of lymphocyte apoptosis in MRL-lpr/lprmice treated with FTY720. Clin. Exp. Immunol 1997, 107, 103–111. [Google Scholar]
- Shalon, D; Smith, SJ; Brown, PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res 1996, 6, 639–645. [Google Scholar]
- Schena, M; Shalon, D; Heller, R; Chai, A; Brown, PO; Davis, RW. Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 1996, 93, 10614–10619. [Google Scholar]
- Paez, GL; Sellers, KF; Band, M; Acland, GM; Zangerl, B; Aguirre, GD. Characterization of gene expression profiles of normal canine retina and brain using a retinal cDNA microarray. Mol. Vis 2006, 12, 1048–1056. [Google Scholar]
- Nagahara, Y; Ikekita, M; Shinomiya, T. Immunosuppressant FTY720 induces apoptosis by direct induction of permeability transition and release of cytochrome c from mitochondria. J. Immunol 2000, 165, 3250–3259. [Google Scholar]
- Fu, NY; Sukumaran, SK; Yu, VC. Inhibition of ubiquitin-mediated degradation of MOAP-1 by apoptotic stimuli promotes Bax function in mitochondria. Proc. Natl. Acad. Sci. USA 2007, 104, 10051–10056. [Google Scholar]
- Tan, KO; Fu, NY; Sukumaran, SK; Chan, SL; Kang, JH; Poon, KL; Chen, BS; Yu, VC. MAP-1 is a mitochondrial effector of Bax. Proc. Natl. Acad. Sci. USA 2005, 102, 14623–14628. [Google Scholar]
- Lee, WJ; Yoo, HS; Suh, PG; Lim, JS; Oh, S; Lee, YM. Sphingosine mediates FTY720-induced apoptosis in LLC-PK1 cells. Exp. Mol. Med 2004, 36, 420–427. [Google Scholar]
- Du, CY; Fang, M; Li, YC; Li, L; Wang, XD. Smac, a mitochondrial protein that promotes cytochrome c–dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar]
- Guo, W; Li, HX. Smac and tumor. Chin. Prac. Med 2008, 3, 192–194. [Google Scholar]
- Zou, H; Henzel, WJ; Liu, XS; Lutschg, A; Wang, XD. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c–dependent activation of caspase-3. Cell 1997, 90, 405–413. [Google Scholar]



| Gene | Sense primer (5′–3′) | Antisense primer (5′–3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| MOAP-1 | GAGTCTCAGGGAACCACGAAG | GGCACAGAAACGACAAAGGGC | 155 | 60 |
| VEGF | CCCACTGAGGAGTCCAACA | CAAATGCTTTCTCCGCTCT | 174 | 57 |
| TRAF6 | CCTGTTGTGATTCATAGCC | GCATCCATTATCTCTTCGTG | 242 | 55 |
| CASP2 | TCCACCCCCACCACTCTTGACT | TGGCTTGCCTTCTTCCCTCTT | 304 | 58 |
| E2F1 | TACCCCAACTCCCTCTACC | TCTGTCTCCCTCCCTCACT | 121 | 62 |
| CASP5 | GTCTAAAGGACAAACCCAAGG | TGTGAAGAGATGAGTGCCAAG | 109 | 57 |
| β–actin | TCATGAAGTGTGACGTGGACATC | TCTAGTTCTAGTAACGAGGAGGAC | 156 | 60 |
| Gene symbol | Gene name | Fold change | P value |
|---|---|---|---|
| Pro-apoptosis genes | |||
| MOAP1 | Modulator of apoptosis 1 | 10.91 | 8.53E-10 |
| VEGF | Vascular endothelial growth factor | 7.72 | 4.01E-05 |
| TRAF6 | TNF receptor-associated factor 6 | 6.93 | 1.31E-05 |
| CASP2 | Caspase 2, apoptosis-related cysteine protease (neural precursor cell expressed, developmentally down regulated 2) | 5.82 | 8.93E-10 |
| E2F1 | E2F transcription factor 1 | 5.68 | 1.25E-06 |
| CASP5 | Caspase 5, apoptosis-related cysteine protease | 5.06 | 1.17E-05 |
| Mdm4 | Mdm4, transformed 3T3 cell double minute 4, p53 binding protein (mouse) | 4.72 | 0.00011 |
| BAX | BCL2-associated X protein | 4.38 | 3.73E-05 |
| BNIP3L | BCL2/adenovirus E1B 19kD interacting protein 3-like | 4.18 | 0.00014 |
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) | 3.64 | 8.12E-05 |
| TUCAN | Tumor up-regulated CARD -containing antagonist of caspase 9 | 3.01 | 8.11E-07 |
| Anti-apoptosis genes | |||
| IGF2 | Insulin-like growth factor 2 (somatomedin A) | 6.17 | 1.81E-05 |
| JUN (AP-1) | V-jun sarcoma virus 17 oncogene homolog (avian) | 4.83 | 0.00017 |
| BIRC1 | Baculoviral IAP repeat-containing 1 | 4.07 | 6.0E-09 |
| FKSG2 | Apoptosis inhibitor | 3.71 | 9.85E-08 |
| BCL2 | B-cell CLL/lymphoma 2 | 0.45 | 0.000377 |
| RAD23B | RAD23 homolog B (S. cerevisiae) | 0.48 | 0.000135 |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, F.; Tan, W.; Guo, D.; Zhu, X.; Qian, K.; He, S. Altered Expression of Signaling Genes in Jurkat Cells upon FTY720 Induced Apoptosis. Int. J. Mol. Sci. 2010, 11, 3087-3105. https://doi.org/10.3390/ijms11093087
Wang F, Tan W, Guo D, Zhu X, Qian K, He S. Altered Expression of Signaling Genes in Jurkat Cells upon FTY720 Induced Apoptosis. International Journal of Molecular Sciences. 2010; 11(9):3087-3105. https://doi.org/10.3390/ijms11093087
Chicago/Turabian StyleWang, Fang, Wenfeng Tan, Dunming Guo, Xiaomin Zhu, Keqing Qian, and Shaoheng He. 2010. "Altered Expression of Signaling Genes in Jurkat Cells upon FTY720 Induced Apoptosis" International Journal of Molecular Sciences 11, no. 9: 3087-3105. https://doi.org/10.3390/ijms11093087




