Molecular Epidemiological Study of Pyrazinamide-Resistance in Clinical Isolates of Mycobacterium tuberculosis from South India
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. Bacterial Samples
3.2. Sputum Processing for AFB Culture
3.3. Drug Susceptibility Test-Proportion Method (Stand and Economic Variant)
3.4. PZase Assay
3.5. Mycobacterium DNA Extraction
3.6. PCR Amplification for Species Identification
3.7. PCR Amplification of pncA
3.8. Agarose Gel Electrophoresis
3.9. Electropherogram Analysis of PCR Amplified Products
3.10. DNA Sequencing
4. Conclusions
Acknowledgments
References and Notes
- Hou, L; Osei Hyiaman, D; Zhang, Z; Wang, B; Yang, A; Kano, K. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol Infect 2000, 124, 227–232. [Google Scholar]
- Barco1, P; Cardoso, RF; Hirata1, RDC; Leite, CQF; Pandolfi, JR; Sato, DN; Shikama, ML; Fiúza de Melo, F; Mamizuka1, EM; Campanerut, PAZ; Hirata, MH. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis clinical isolates from the southeast region of Brazil. J. Antimicrob. Chemother 2006, 58, 930–935. [Google Scholar]
- Boshoff, HI; Mizrahi, V. Expression of Mycobacterium smegmatis pyrazinamidase in Mycobacterium tuberculosis confers hypersensitivity to pyrazinamide and related amides. J. Bacteriol 2000, 182, 5479–5485. [Google Scholar]
- Boshoff, HI; Mizrahi, V; Barry, CE. Effects of pyrazinamide on fatty acid synthesis by whole mycobacterial cells and purified fatty acid synthase. J. Bacteriol 2002, 184, 2167–2172. [Google Scholar]
- Huang, T; Lee, SS; Tu, H. Correlation between Pyrazinamide activity and pncA mutations in Mycobacterium tuberculosis isolates in Taiwan. Antimicrob. Agents Chemother 2003, 47, 3672–3673. [Google Scholar]
- Portugal, I; Barreiro, L; Moniz-Pereira, J. pncA mutations in Pyrazinamide-Resistant Mycobacterium tuberculosis Isolates in Portugal. Antimicrob. Agents Chemother 2004, 48, 2736–2768. [Google Scholar]
- Hewlett, D; Horn, DL; Alfalla, C. Drug-resistant tuberculosis: Inconsistent results of pyrazinamide susceptibility testing. JAMA 1995, 12, 916–917. [Google Scholar]
- Raynaud, C; Lane elle, M; Senaratne, RH. Mechanisms of pyrazinamide resistance in mycobacteria: Importance of lack of uptake in addition to lack of pyrazinamidase activity. Microbiology 1999, 145, 1359–1367. [Google Scholar]
- Wade, MM; Volokhov, D; Peredelchuk, M. Accurate mapping of mutations of pyrazinamide-resistant Mycobacterium tuberculosis strains with a scanning-frame oligonucleotide microarray. Diagn. Microbiol. Infect. Dis 2004, 49, 89–97. [Google Scholar]
- Zhang, Y; Scorpio, A; Nikaido, H. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of M. tuberculosis to pyrazinamide. J. Bacteriol 1999, 181, 2044–2049. [Google Scholar]
- Park, SK; Lee, J Yoo; Chang, CL; Lee, MK; Son, HC; Kim, CM; Jang, HJ; Park, HK; Jeong, SH. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect. Dis 2001, 1, 4. [Google Scholar]
- Scorpio, A; Lindholm-Levy, P; Heifets, L. Characterization of pnc A mutations in Pyrazinamide resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother 1997, 41, 540–543. [Google Scholar]
- Sreevatsan, S; Pan, X; Zhang, Y; Kreiswirth, BN; Musser, JM. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob. Agents Chemother 1997, 41, 636–640. [Google Scholar]
- Hirano, K; Takahashi, M; Kazumi, Y; Fukasawa, Y; Abe, C. Mutation in pncA is a major mechanism of Pyrazinamide resistance in Mycobacterium tuberculosis. Tubercle. Lung Dis 1998, 78, 117–122. [Google Scholar]
- Mestdagh, M; Fonteyne, PA; Realini, L. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother 1999, 43, 2317–2319. [Google Scholar]
- Hou, L; Osei-Hyiaman, D; Zhang, Z. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol. Infect 2000, 124, 227–32. [Google Scholar]
- Lemaitre, N; Sougakoff, W; Truffot-Pernot, C. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase pncA. Antimicrob. Agents Chemother 1999, 43, 1761–1763. [Google Scholar]
- Scorpio, A; Lindholm-Levy, P; Heifets, L. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother 1997, 41, 540–543. [Google Scholar]
- Park, SK; Lee, JY; Chang, CL. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect. Dis 2001, 1, 4. [Google Scholar]
- Rodrigues, VFS; Telles, MA; Ribeiro, MO. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis in Brazil. Antimicrob. Agents Chemother 2005, 49, 444–446. [Google Scholar]
- Heifets, L; Lindholm-Levy, P. Pyrazinamide sterilizing activity in vitro against semi-dormant Mycobacterium tuberculosis bacterial populations. Am. Rev. Respir. Dis 1992, 145, 1223–1225. [Google Scholar]
- Davies, AP; Billington, OJ; McHugh, TD; Mitchison, DA; Gillespie, SH. Comparison of phenotypic and genotypic methods for Pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol 2000, 38, 3686–3688. [Google Scholar]
- Morlock, GP; Crawford, JT; Butler, WR; Brim, SE; Sikes, D; Mazurek, GH; Woodley, CL; Cooksey, RC. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother 2000, 44, 2291–2295. [Google Scholar]
- Hirano, K; Takahashi, M; Kazumi, Y; Fukasawa, Y; Abe, C. Mutation in pncA is a major mechanism of Pyrazinamide resistance in Mycobacterium tuberculosis. Tubercle. Lung. Dis 1998, 78, 117–122. [Google Scholar]
- Isenberg, HD. Mycobacteriology: Identification tests for mycobacteria. In Clinical Microbiology Procedure Handbook; Isenberg, HD, Ed.; ASM Press: Washington, DC, USA, 1995; Volume 1, pp. 12–17. [Google Scholar]
- Mani, C; Selvakumar, N; Kumar, V; Narayanan, S; Narayanan, PR. Comparison of DNA sequencing, PCR-SSCP and PhaB assays with indirect sensitivity testing for detection of rifampcin resistance in Mycobacterium tuberculosis. Int. J. Tuber. Lung. Dis 2003, 7, 625–659. [Google Scholar]
- Fletcher, HA; Donoghue, HD; Taylor, GM; Adri, GM; Van der Zanden, AGM; Spigelman, M. Molecular analysis of Mycobacterium tuberculosis DNA from a family of 18th century Hungarians. Microbiology 2003, 149, 143–151. [Google Scholar]
- Scorpio, A; Lindholm-levy, P; Heifets, L; Gilman, R; Siddiqi, S; Cynamon, M; Zhang, Y. Characterization of pncA Mutations in Pyrazinamide-Resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother 1997, 41, 540–543. [Google Scholar]
- Hathaway, LJ; Brugger, S; Martynova, A; Aebi, S; Mu¨hlemann, K. Use of the Agilent 2100 Bioanalyzer for Rapid and Reproducible Molecular Typing of Streptococcus pneumoniae. J. Clin. Microbiol 2007, 45, 803–809. [Google Scholar]
- Altschul, SF; Madden, TL; Schaffer, AA; Zhang, J; Zhang, Z; Miller, W; Lipman, DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. J. Nucleic. Acids Res 1997, 25, 3389–3402. [Google Scholar]
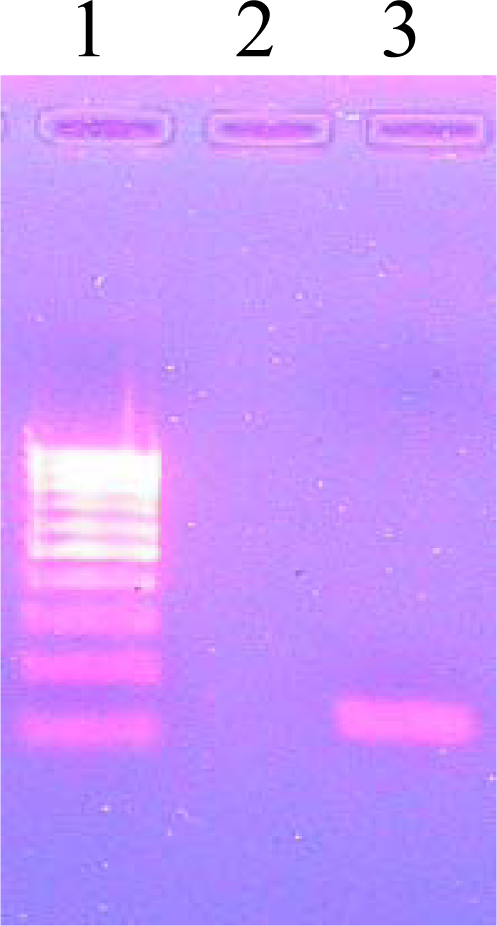
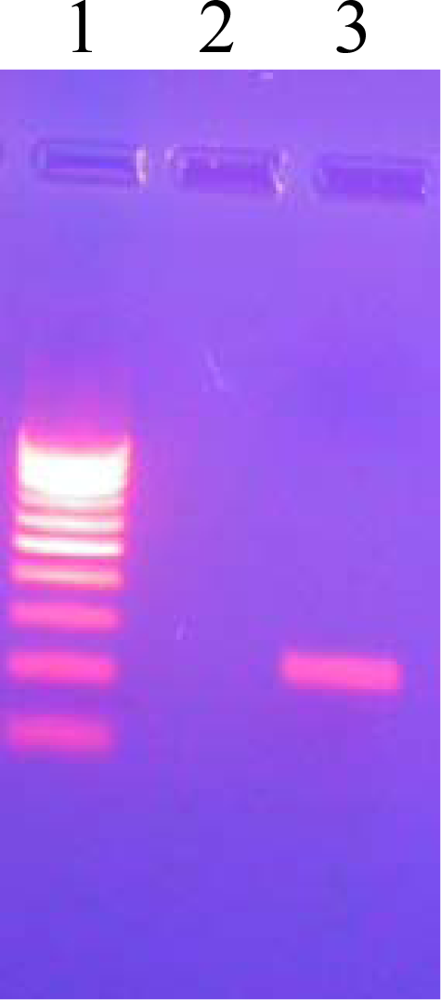
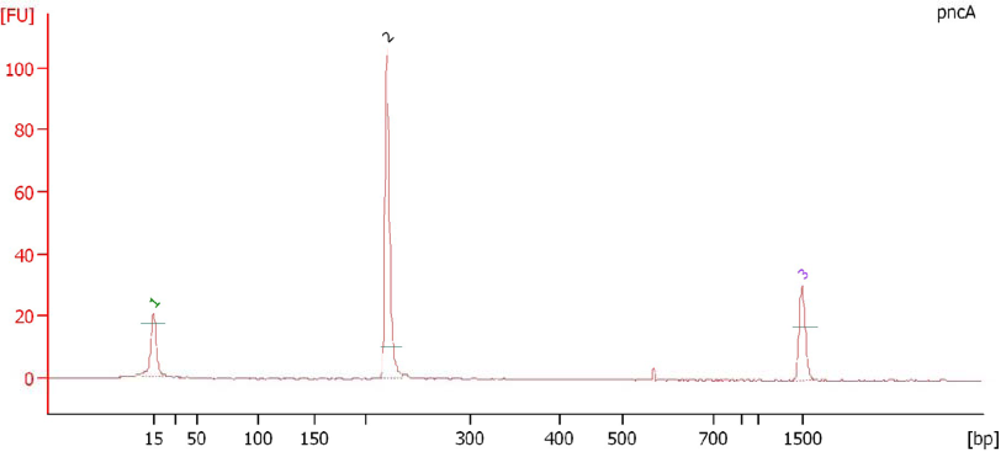
| PCR Mediated Direct sequencing | In vitro sensitivity tests on L.J. slants (μg/mL) | |||
|---|---|---|---|---|
| Susceptibility (MIC = 100) | Resistance | PZase activity | ||
| MIC = 300 | MIC > 300 | |||
| Resistant 39 (78%) | 00 | 26(52%) | 13(26%) | Negative 39 (78%) |
| Sensitive 11 (22%) | 11(22%) | -- | -- | Positive 11 (22%) |
| Total 50 (100%) | 11(22%) | 39(78%) | Total 50 (100%) | |
| Mutation Site | Nucleotide Changes | Amino Acid Changes | No. of Isolates | Percentage (%) |
|---|---|---|---|---|
| 137 | A→C | His→Pro | 2 | 5.1 |
| 138 | G→A | Cys→Tyr | 2 | 5.1 |
| 139 | G→C | Val→Leu | 2 | 5.1 |
| 142 | C→T | Thr→Met | 2 | 5.1 |
| 5 | AT insertion | Ile→Ser | 2 | 5.1 |
| 12 | A→C | Asp→Ala | 4 | 10.3 |
| 26 | G→C | Ala→Gly | 2 | 5.1 |
| 51 | C→G | His→Gln | 2 | 5.1 |
| 69 | C→G | Pro→Arg | 3 | 7.7 |
| 72 | T→C | Cys→Arg | 3 | 7.7 |
| 85 | T→C | Leu→Pro | 6 | 15.4 |
| 96 | A→C | Lys →Asn | 37.7 | |
| 132 | TC insertion | Gly→Ser | 3 | 7.7 |
| 141 | C→A | Gln→Pro | 1 | 2.6 |
| 142 | AG insertion | Thr→Lys | 1 | 2.6 |
| 171 | G→C | Ala→Pro | 1 | 2.6 |
| Total | 39 | 100 |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Muthaiah, M.; Jagadeesan, S.; Ayalusamy, N.; Sreenivasan, M.; Prabhu, S.S.; Muthuraj, U.; Senthilkumar, K.; Veerappan, S. Molecular Epidemiological Study of Pyrazinamide-Resistance in Clinical Isolates of Mycobacterium tuberculosis from South India. Int. J. Mol. Sci. 2010, 11, 2670-2680. https://doi.org/10.3390/ijms11072670
Muthaiah M, Jagadeesan S, Ayalusamy N, Sreenivasan M, Prabhu SS, Muthuraj U, Senthilkumar K, Veerappan S. Molecular Epidemiological Study of Pyrazinamide-Resistance in Clinical Isolates of Mycobacterium tuberculosis from South India. International Journal of Molecular Sciences. 2010; 11(7):2670-2680. https://doi.org/10.3390/ijms11072670
Chicago/Turabian StyleMuthaiah, Muthuraj, Sridharan Jagadeesan, Nisha Ayalusamy, Manupriya Sreenivasan, Sambamurthy Sangamesvara Prabhu, Usharani Muthuraj, Kamatchiyammal Senthilkumar, and Saroja Veerappan. 2010. "Molecular Epidemiological Study of Pyrazinamide-Resistance in Clinical Isolates of Mycobacterium tuberculosis from South India" International Journal of Molecular Sciences 11, no. 7: 2670-2680. https://doi.org/10.3390/ijms11072670




