Over-expression, Rapid Preparation and Some Properties of C-terminal BARc Region in PICK1
Abstract
:1. Introduction
2. Materials and methods
2.1. Bacterial strains, plasmid and reagents
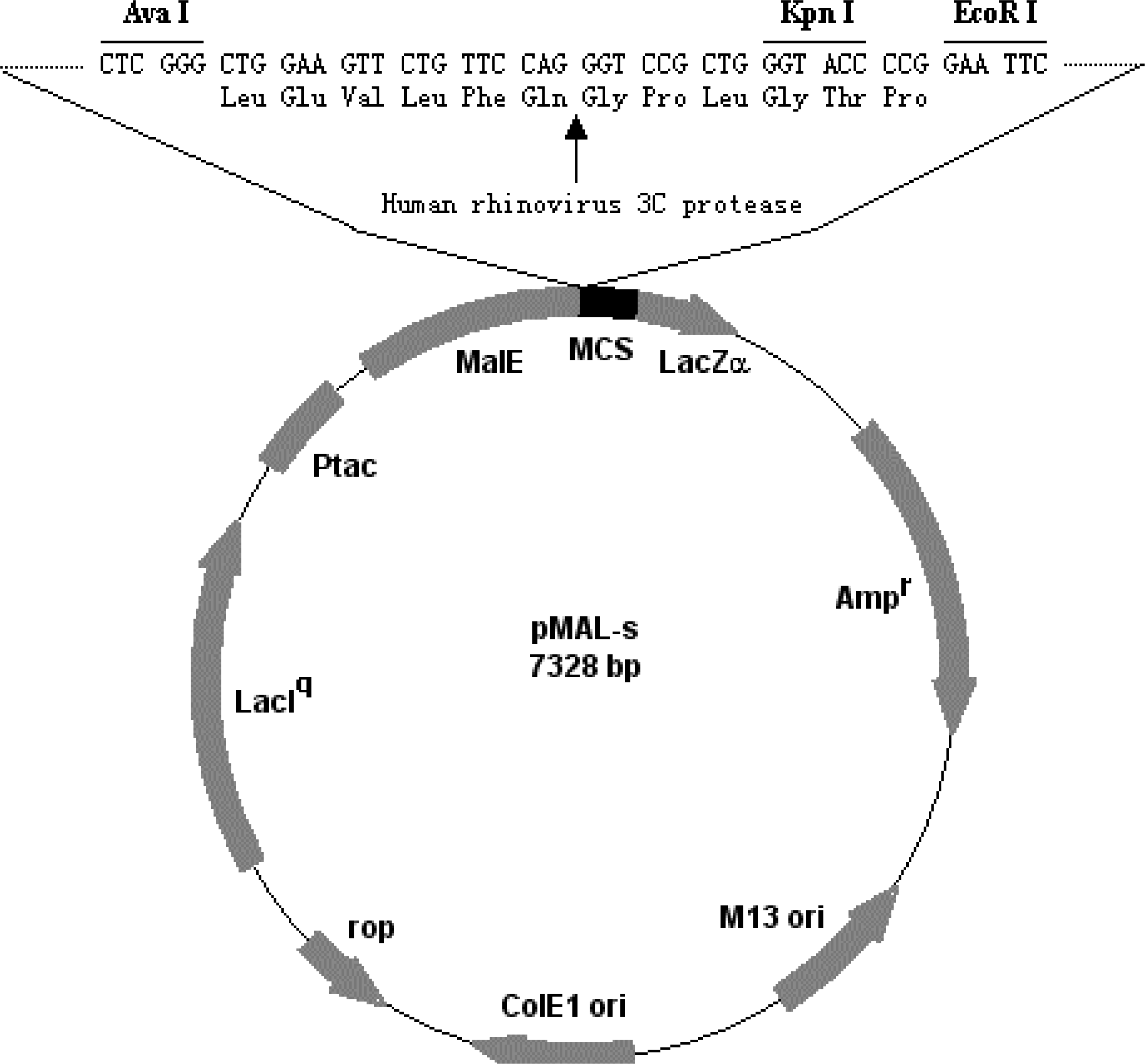
2.2. Construction of recombinant plasmid pMAL-s-barc
2.3. Expression and purification of fusion protein, MBP-BARc
2.4. Removal of the affinity tag and separation of the BARc
2.5. Western-blot analysis of BARc domain
2.6. Interaction of PDZ and BARc domains identified by the Pull-down test
2.7. Protein Lipid Overlay (PLO) assay
3. Results and Discussion
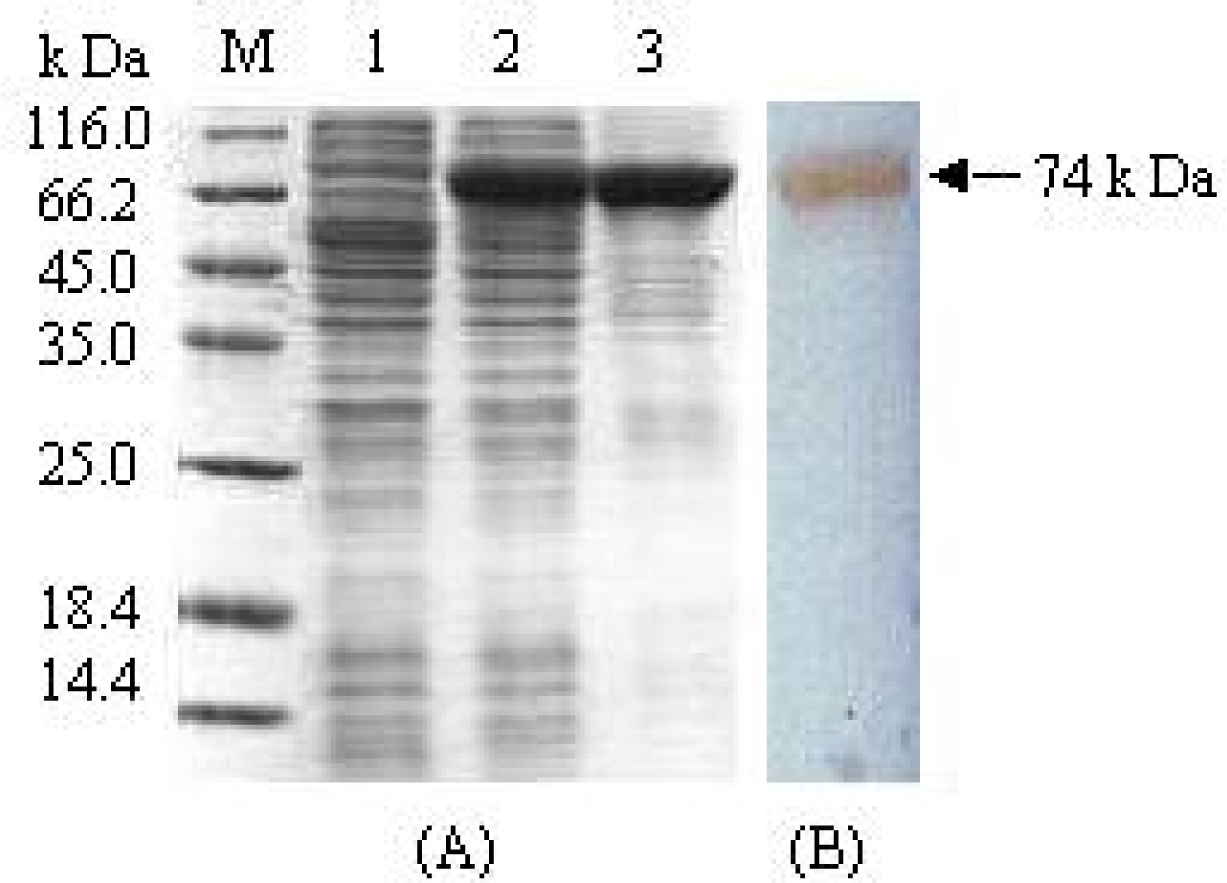
3.1. Expression and purification of fusion protein MBP-BARc
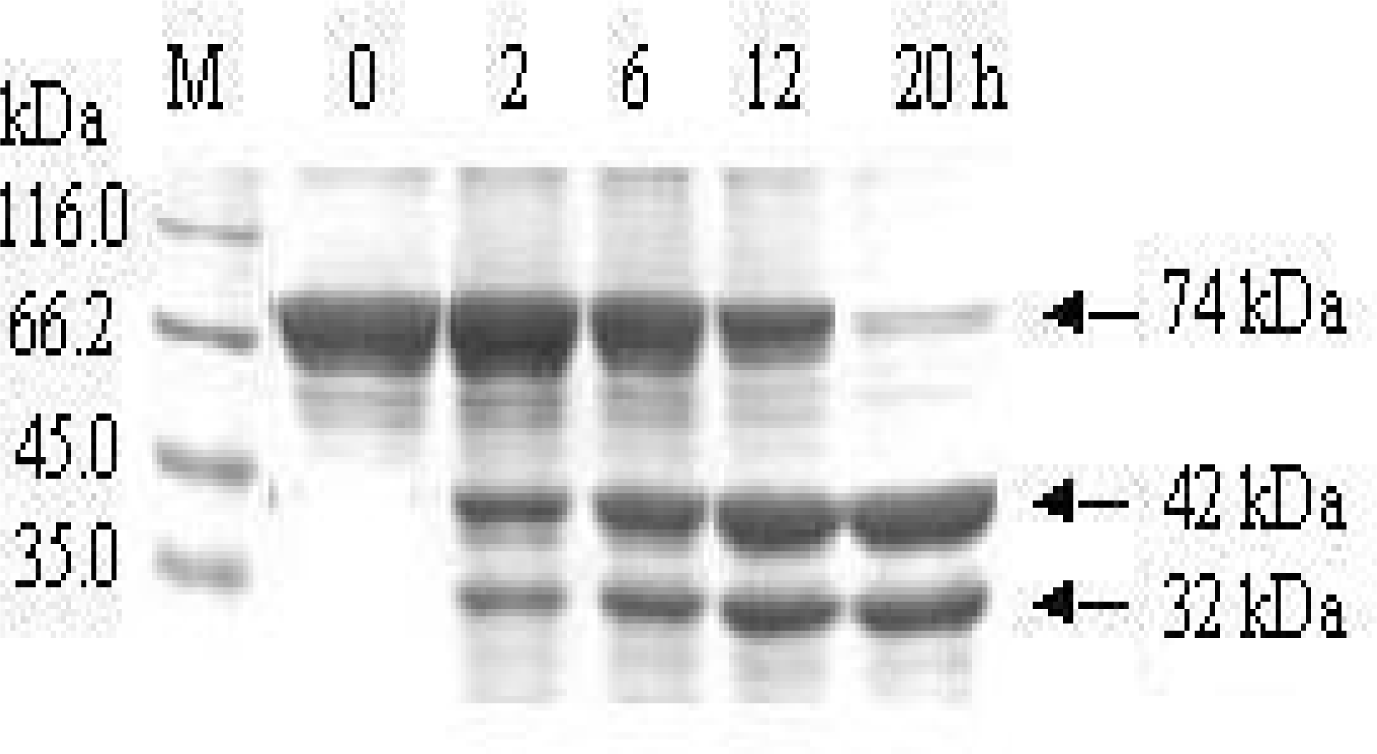
3.2. Removal of the affinity tag from MBP-BARc
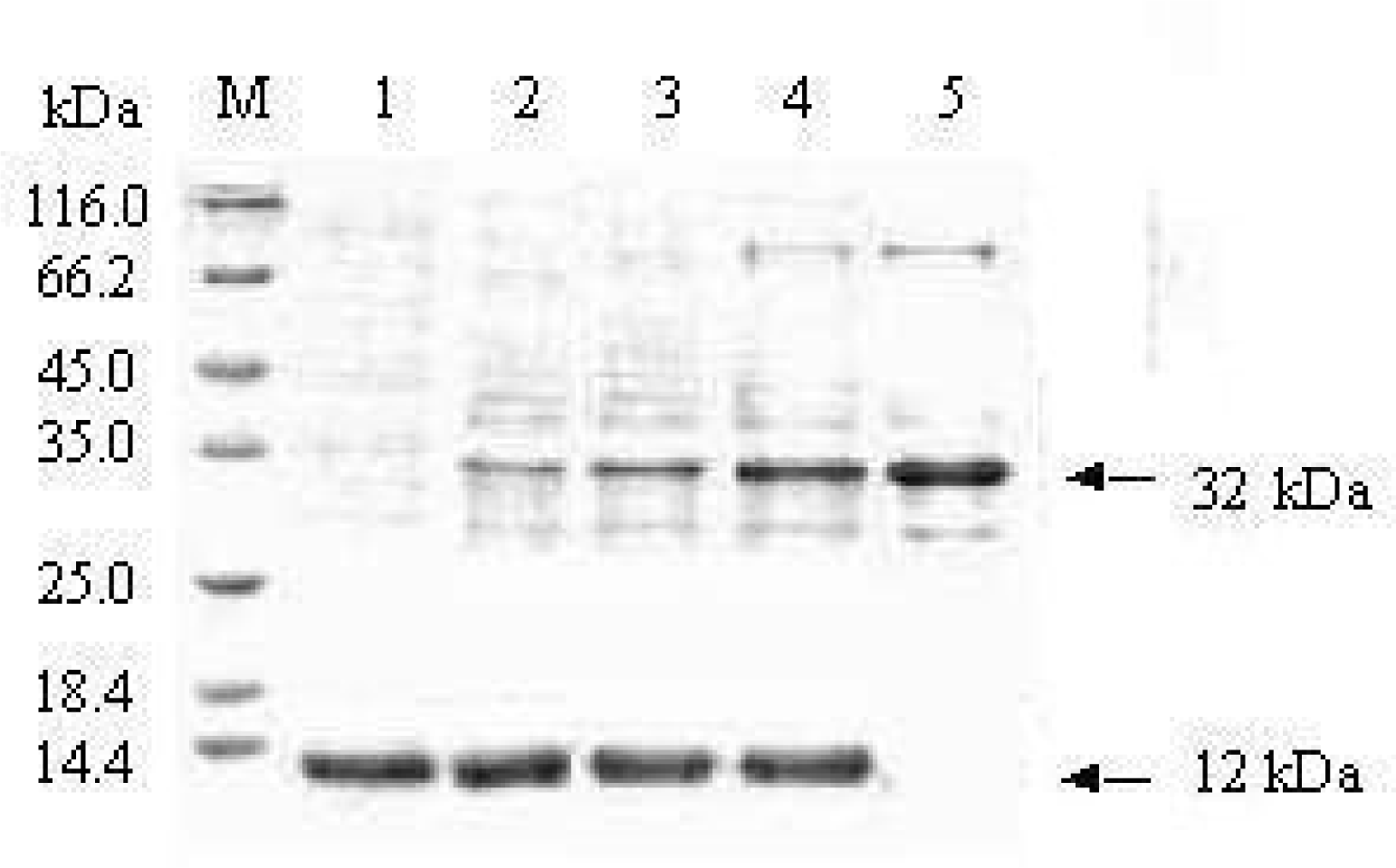
3.4. Protein-protein interaction identified by Pull-down test
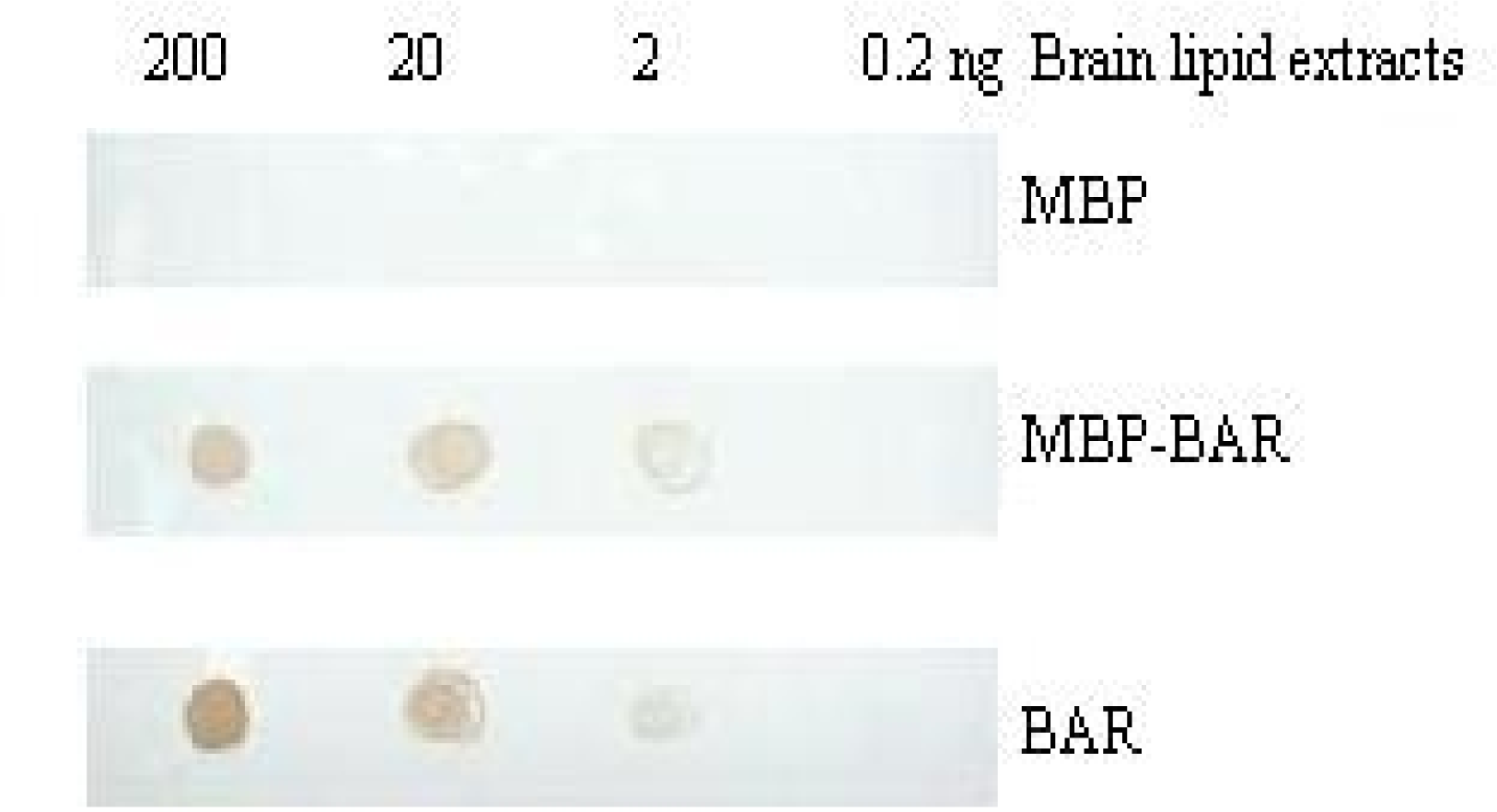
3.5. Combination of the BARc with lipid by PLO assay
4. Conclusions
Acknowledgments
References
- Staudinger, J; Zhou, J; Burgess, R; Elledge, SJ; Olson, EN. PICK1: A perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol 1995, 128, 263–271. [Google Scholar]
- Madsen, KL; Beuming, T; Niv, MY; Chang, CW; Dev, KK; Weinstein, H; Gether, U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem 2005, 280, 20539–20548. [Google Scholar]
- Hanley, JG; Khatri, L; Hanson, PI; Ziff, EB. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron 2002, 34, 53–67. [Google Scholar]
- Xu, J; Xia, J. Structure and function of PICK1. Neurosignals 2007, 15, 190–201. [Google Scholar]
- Dev, KK. PDZ domain protein-protein interactions: a case study with PICK1. Curr. Top Med. Chem 2007, 7, 3–20. [Google Scholar]
- Peter, BJ; Kent, HM; Mills, IG; Vallis, Y; Butler, PJ; Evans, PR; McMahon, HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar]
- Habermann, B. The BAR-domain family of proteins: a case of bending and binding? EMBO J 2004, 5, 250–255. [Google Scholar]
- Masuda, M; Takeda, S; Sone, M; Ohki, T; Mori, H; Kamioka, Y; Mochizuki, N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J 2006, 25, 2889–2897. [Google Scholar]
- Xiao, H; Shi, YW; Wang, LL; Yuan, JM. Protein-protein interaction between domains of PDZ and BAR from PICK1. Chem. Res. Chin. U 2007, 23, 191–195. [Google Scholar]
- Li, HJ; Wang, WJ; Shi, R. Gene cloning, expression, purification and activity of human rhinovirus 3C protease. Chin. J. Biolocals 2007, 20, 240–243. [Google Scholar]
- Li, ZY; Li, YJ; Guo, CY; Shi, YW; Xu, MQ; Trommer, WE; Yuan, JM. Soluble expression and affinity purification of functional domain of human acetylcholine receptor α-subunit by the modulation of maltose binding protein. Biotechnol. Lett 2004, 26, 1765–1769. [Google Scholar]
- Singh, D; Lampe, PD. Identification of connexin-43 interacting proteins. Cell Commun. Adhes 2003, 10, 215–220. [Google Scholar]
- Dowler, S; Kular, G; Alessi, DR. Protein lipid overlay assay. Sci. STKE 2002, 129, PL6. [Google Scholar]
- Jin, W; Ge, WP; Xu, J; Cao, M; Peng, L; Yung, W; Liao, D; Duan, S; Zhang, M; Xia, J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci 2006, 26, 2380–2390. [Google Scholar]

© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/). This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xiao, H.; Shi, Y.; Yuan, J.; Huang, Y.; Wang, J. Over-expression, Rapid Preparation and Some Properties of C-terminal BARc Region in PICK1. Int. J. Mol. Sci. 2009, 10, 28-36. https://doi.org/10.3390/ijms10010028
Xiao H, Shi Y, Yuan J, Huang Y, Wang J. Over-expression, Rapid Preparation and Some Properties of C-terminal BARc Region in PICK1. International Journal of Molecular Sciences. 2009; 10(1):28-36. https://doi.org/10.3390/ijms10010028
Chicago/Turabian StyleXiao, Hong, Yawei Shi, Jingming Yuan, Yuming Huang, and Junhua Wang. 2009. "Over-expression, Rapid Preparation and Some Properties of C-terminal BARc Region in PICK1" International Journal of Molecular Sciences 10, no. 1: 28-36. https://doi.org/10.3390/ijms10010028



