The Effects of Berberine on the Gut Microbiota in Apc min/+ Mice Fed with a High Fat Diet
Abstract
:1. Introduction
2. Results
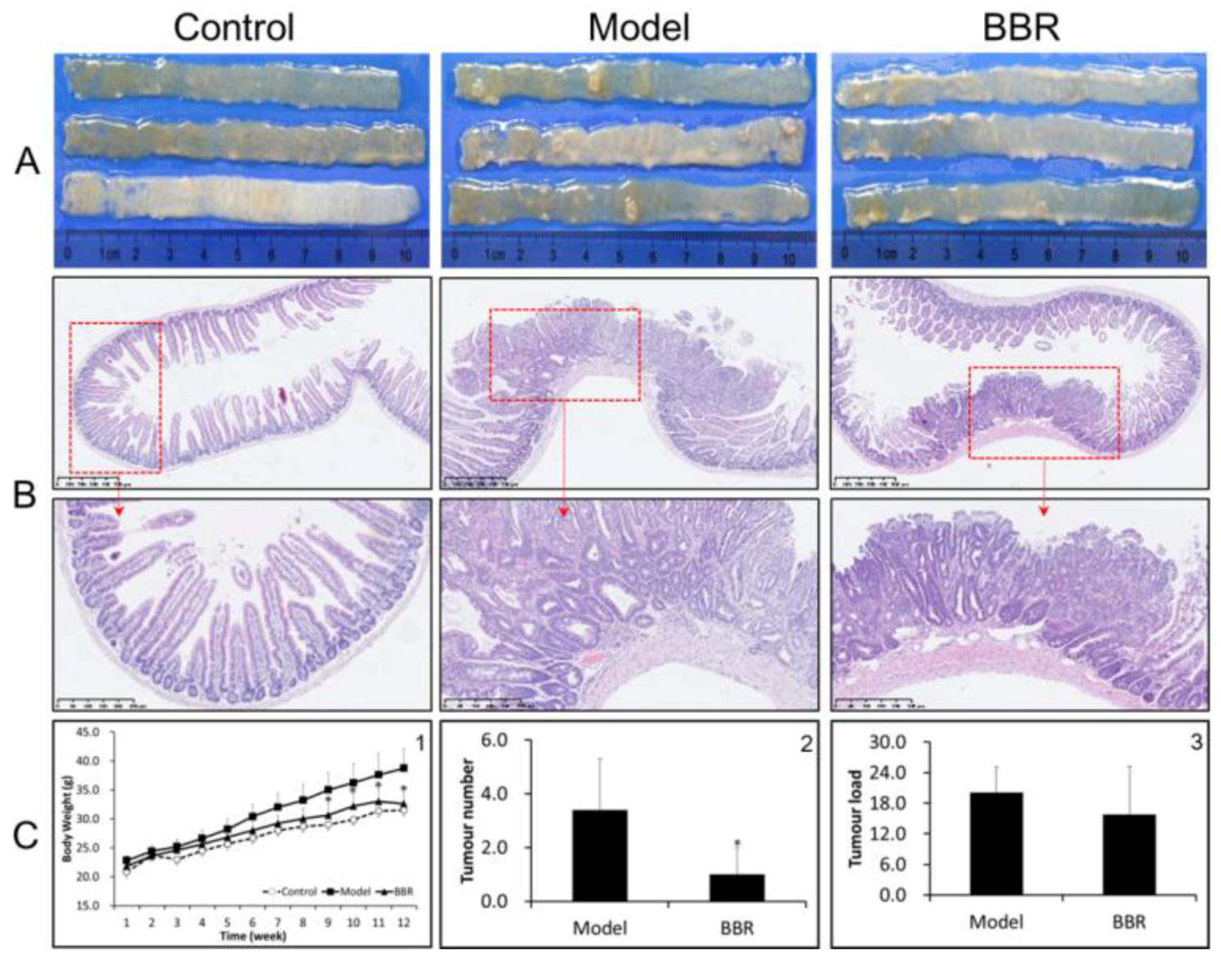
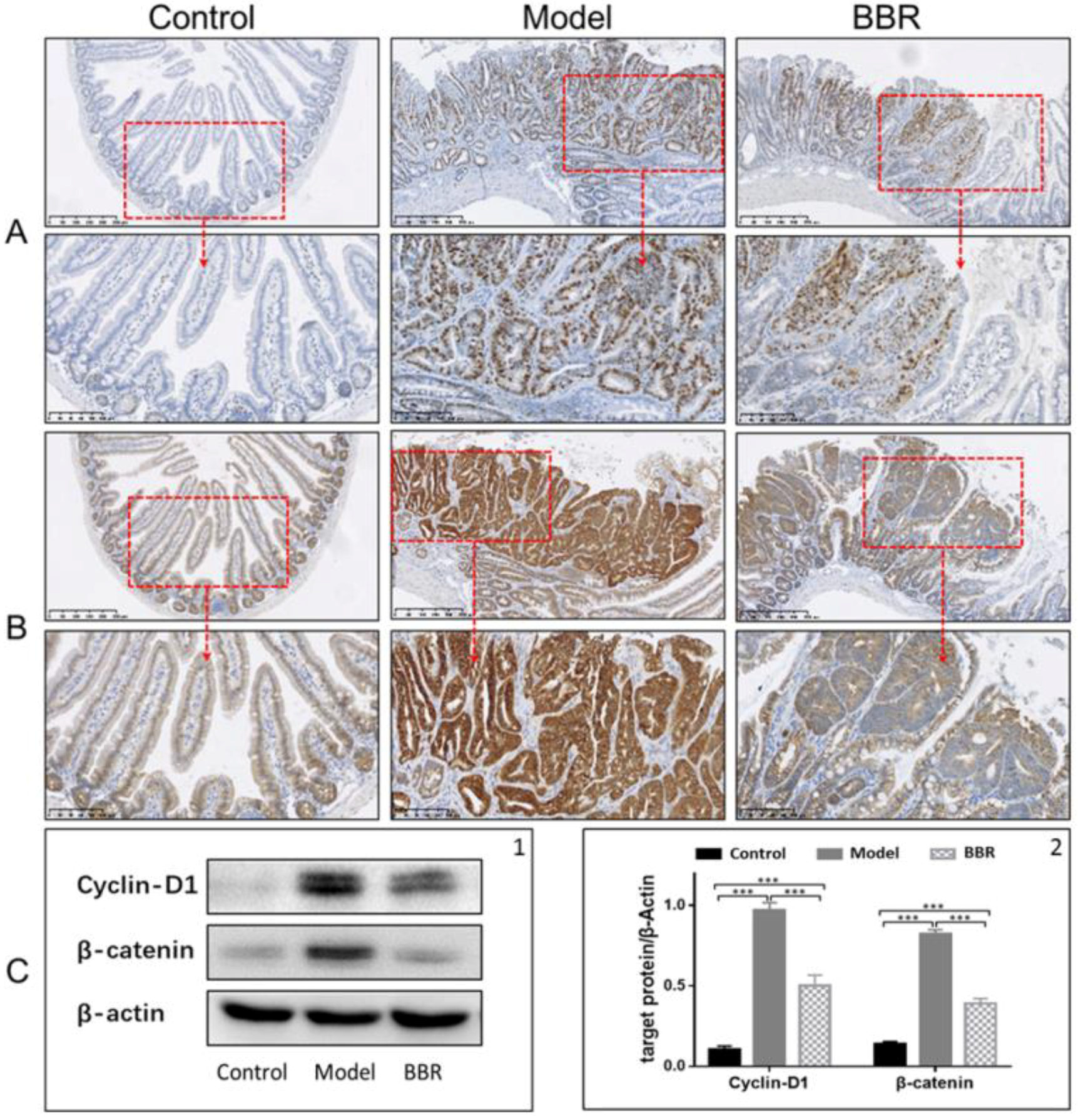
2.1. Effects of BBR on Intestinal Adenoma Development and β-Catenin Signaling in Apc min/+ Mice Fed with a HFD
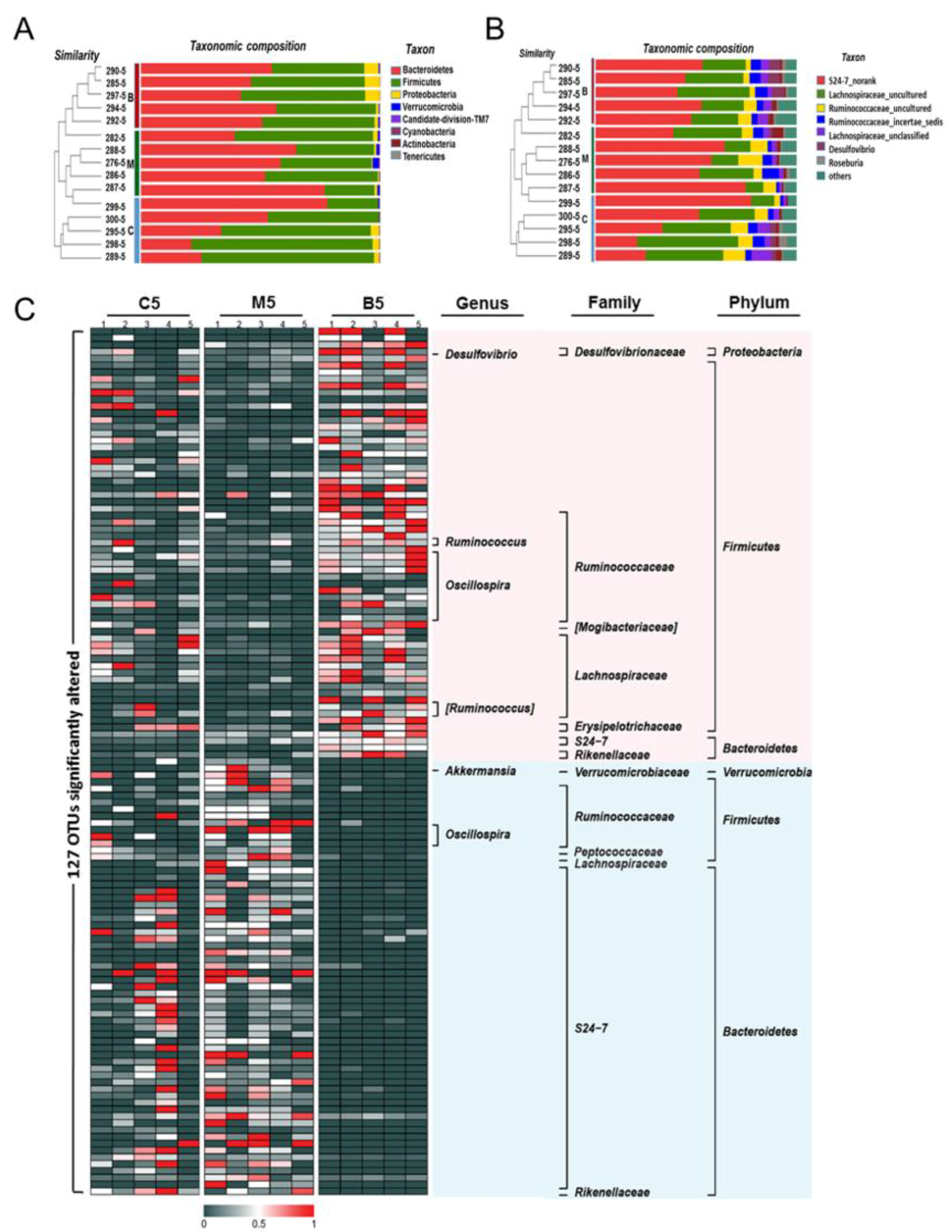
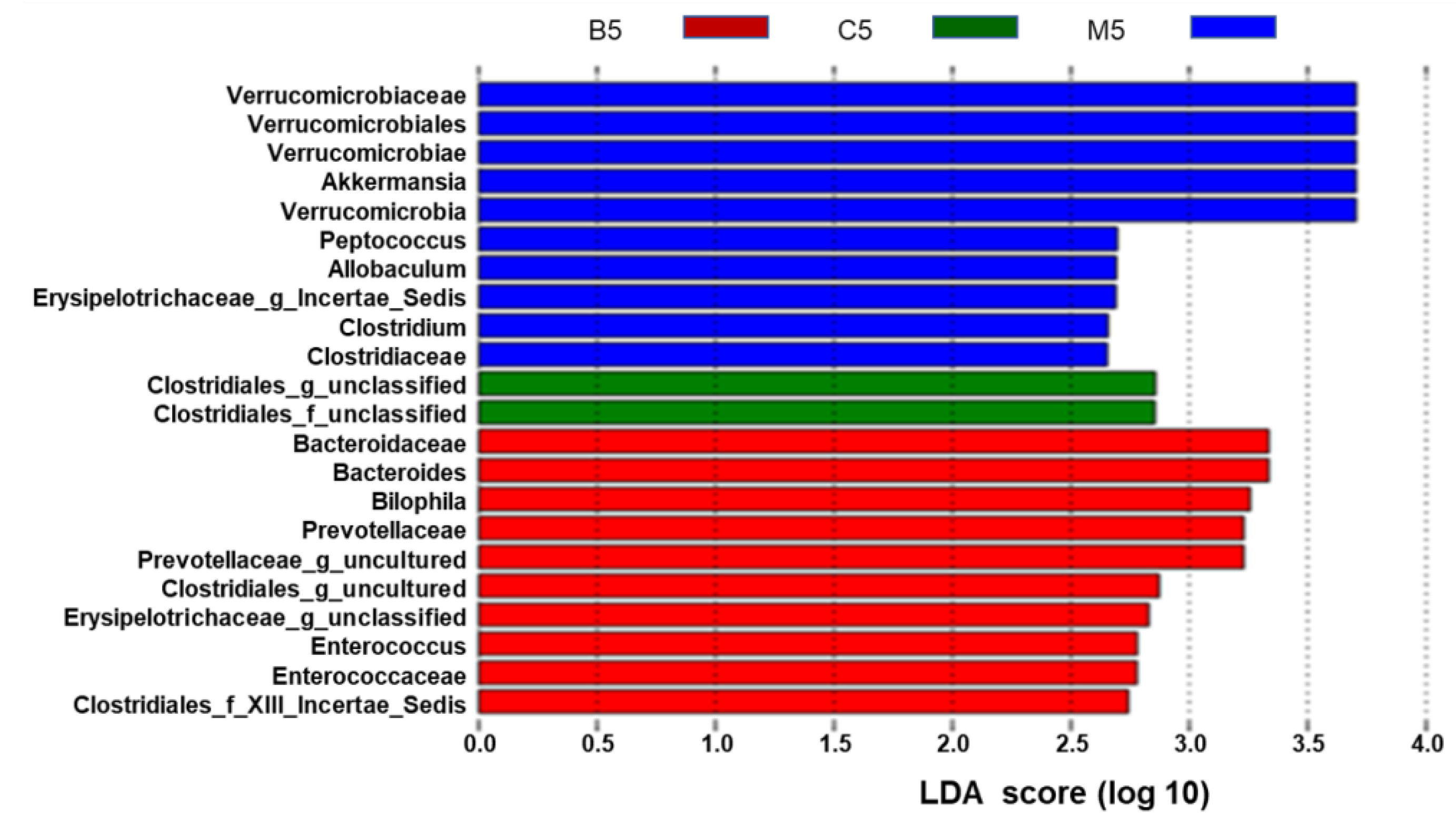
2.2. Overall Effects of BBR on the Gut Microbiota Structure in Apc min/+ Mice Fed with an HFD
2.3. Effects of BBR on Gut Microbiota in Apc min/+ Mice Fed with an HFD at Different Levels
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animals and Experimental Design
4.3. Histological Examination
4.4. Immunohistochemistry
4.5. Western Blotting Analysis
4.6. DNA Extraction, PCR Amplification, and Illumina MiSeq Sequencing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Huang, C.; Wu, L.; Wen, B. Effect of evodiamine and berberine on miR-429 as an oncogene in human colorectal cancer. Onco Targets Ther. 2016, 9, 4121–4127. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Pan, K.; Linnekamp, J.F.; Medema, J.P.; Kandimalla, R. DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta 2016, 1866, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huycke, M.M. Colorectal cancer: Role of commensal bacteria and bystander effects. Gut Microbes 2015, 6, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Bultman, S.J. The microbiome and its potential as a cancer preventive intervention. Semin. Oncol. 2016, 43, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.; Pan, W.; Cai, Y. Gut microbiota and colorectal cancer: Insights into pathogenesis for novel therapeutic strategies. Gastroenterol. 2017, 55, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Vallance, B.A.; Boyer, L.; Bergstrom, K.S.; Walker, J.; Madsen, K.; O’Kusky, J.R.; Buchan, A.M.; Jacobson, K. Saccharomyces boulardii ameliorates Citrobacterrodentium-induced colitis through actions on bacterial virulence factors. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Z.; Yu, C.; Wen, X.D.; Chen, L.; Zhang, C.F.; Calway, T.; Qiu, Y.; Wang, Y.; Zhang, Z.; Anderson, S.; et al. American Ginseng Attenuates Colitis-Associated Colon Carcinogenesis in Mice: Impact on Gut Microbiota and Metabolomics. Cancer Prev. Res. (Phila) 2016, 9, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, L.; Chang, E.B.; Wang, J.Y.; Raufman, J.P. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol. Cancer 2015, 14, 180. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Y.; Huycke, M.M. Microbiome-driven carcinogenesis in colorectal cancer: Models and mechanisms. Free Radical. Biol. Med. 2017, 105, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Zhang, M.; Pang, X.; Xu, J.; Kang, C.; Li, M.; Zhang, C.; Zhang, Z.; Zhang, Y.; et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, Y.; Huang, W.; Selwyn, F.P.; Klaassen, C.D. Dose-response effect of berberine on bile acid profile and gut microbiota in mice. BMC Complement. Altern. Med. 2016, 16, 394. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S. Berberine and inflammatory bowel disease: A concise review. Pharmacol. Res. 2016, 113, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, Y.; Liu, K.; Zhao, Y.; Xu, B.; Xu, L.; Tan, L.; Tian, Y.; Li, C.; Zhang, W.; et al. Berberine inhibits colitis-associated tumorigenesis via suppressing inflammatory responses and the consequent EGFR signaling-involved tumor cell growth. Lab. Investig. 2017, 97, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hua, B.; Saud, S.M.; Lin, H.; Hou, W.; Matter, M.S.; Jia, L.; Colburn, N.H.; Young, M.R. Berberine regulates AMP-activated protein kinase signaling pathways and inhibits colon tumorigenesis in mice. Mol. Carcinog. 2015, 54, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.M.; Ming, Z.; Shen, G.W.; Xiao, J.B.; Hatch, G.M.; Jing, K.G.; Li, C. Amorphous solid dispersion of berberine with absorption enhancer demonstrates a remarkable hypoglycemic effect via improving its bioavailability. Int. J. Pharm. 2014, 467, 50–59. [Google Scholar] [CrossRef]
- Sun, R.; Yang, N.; Kong, B.; Cao, B.; Feng, D.; Yu, X.; Ge, C.; Huang, J.; Shen, J.; Wang, P.; et al. Orally Administered Berberine Modulates Hepatic Lipid Metabolism by Altering Microbial Bile Acid Metabolism and the Intestinal FXR Signaling Pathway. Mol. Pharmacol. 2017, 91, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.H.; Liu, X.Z.; Pan, W.; Zou, D.J. Berberine protects against diet-induced obesity through regulating metabolic endotoxemia and gut hormone levels. Mol. Med. Rep. 2017, 15, 2765–2787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Pan, Q.; Cai, W.; Shen, F.; Chen, G.Y.; Xu, L.M.; Fan, J.G. Modulation of Gut Microbiota by Berberine Improves Steatohepatitis in High-Fat Diet-Fed BALB/C. Mice. Arch. Iran. Med. 2016, 19, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Fukugaiti, M.H.; Ignacio, A.; Fernandes, M.R.; Ribeiro, J.U.; Nakano, V.; Avila-Campos, M.J. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz. J. Microbiol. 2015, 46, 1135–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chidambara, M.K.N.; Jayaprakasha, G.K.; Patil, B.S. The natural alkaloid berberine targets multiple pathways to induce cell death in cultured human colon cancer cells. Eur. J. Pharmacol. 2012, 688, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, H.; Zhang, B.; Cao, H.; Xu, X.; Ruan, H.; Yi, T.; Tan, L.; Qu, R.; Song, G.; et al. Berberine potently attenuates intestinal polyps growth in ApcMin mice and familial adenomatous polyposis patients through inhibition of Wnt signalling. J. Cell Mol. Med. 2013, 17, 1484–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calle, E.E.; Thun, M.J. Obesity and cancer. Oncogene 2004, 23, 6365–6378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Edelmann, W.; Fan, K.; Lau, K.; Leung, D.; Newmark, H.; Kucherlapati, R.; Lipkin, M. Dietary modulation of carcinoma development in a mouse model for human familial adenomatous polyposis. Cancer Res. 1998, 58, 5713–5717. [Google Scholar] [PubMed]
- Talla, S.B.; Brembeck, F.H. The role of Pygo2 for Wnt/β-catenin signaling activity during intestinal tumor initiation and progression. Oncotarget 2016, 7, 80612–80632. [Google Scholar] [CrossRef] [PubMed]
- Nagalingam, N.A.; Kao, J.Y.; Young, V.B. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 2011, 17, 917–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucke, K.; Miehlke, S.; Jacobs, E.; Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006, 55, 617–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Wu, Y.; Deng, B.; Li, J.; Cao, H.; Qu, Y.; Qian, X.; Zhong, G. Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget 2016, 7, 85318–85331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, N.T.; Zackular, J.P.; Chen, G.Y.; Schloss, P.D. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014, 2, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zackular, J.P.; Rogers, M.A.; Ruffin, M.T. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. (Phila) 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Jin, Z.; Wu, W.; Gao, R.; Guo, B.; Gao, Z.; Yang, Y.; Qin, H. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS ONE 2014, 9, e90849. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.D.; Wang, C.Z.; Yu, C.; Zhao, L.; Zhang, Z.; Matin, A.; Wang, Y.; Li, P.; Xiao, S.Y.; Du, W.; et al. Panax noto ginseng attenuates experimental colitis in the azoxymethane/dextran sulfate sodium mouse model. Phytother. Res. 2014, 28, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, Z.M.; Liu, Q.; Li, Y.; Ma, Y.; Wang, J.; Sun, M.R.; Shao, H.B.; Sun, H.; Malin, G.; et al. Comparative study of the composition and genetic diversity of the picoeukaryote community in a Chinese aquaculture area and an open sea area. J. Mar. Biol. Assoc. UK 2016, 97, 151–159. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds BBR are available from the authors. |

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Guan, L.; Li, J.; Lai, M.; Wen, X. The Effects of Berberine on the Gut Microbiota in Apc min/+ Mice Fed with a High Fat Diet. Molecules 2018, 23, 2298. https://doi.org/10.3390/molecules23092298
Wang H, Guan L, Li J, Lai M, Wen X. The Effects of Berberine on the Gut Microbiota in Apc min/+ Mice Fed with a High Fat Diet. Molecules. 2018; 23(9):2298. https://doi.org/10.3390/molecules23092298
Chicago/Turabian StyleWang, Huan, Lingnan Guan, Jing Li, Maode Lai, and Xiaodong Wen. 2018. "The Effects of Berberine on the Gut Microbiota in Apc min/+ Mice Fed with a High Fat Diet" Molecules 23, no. 9: 2298. https://doi.org/10.3390/molecules23092298