Nitrate Accumulation and Expression Patterns of Genes Involved in Nitrate Transport and Assimilation in Spinach
Abstract
:1. Introduction
2. Results
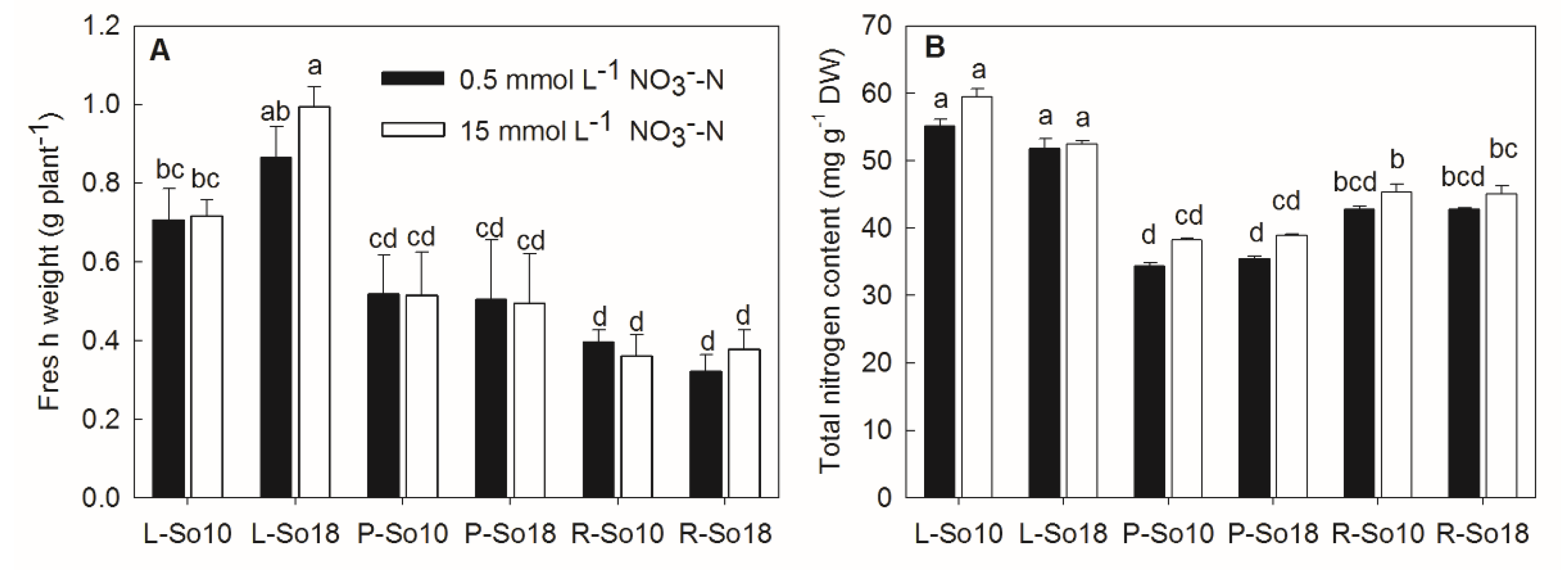
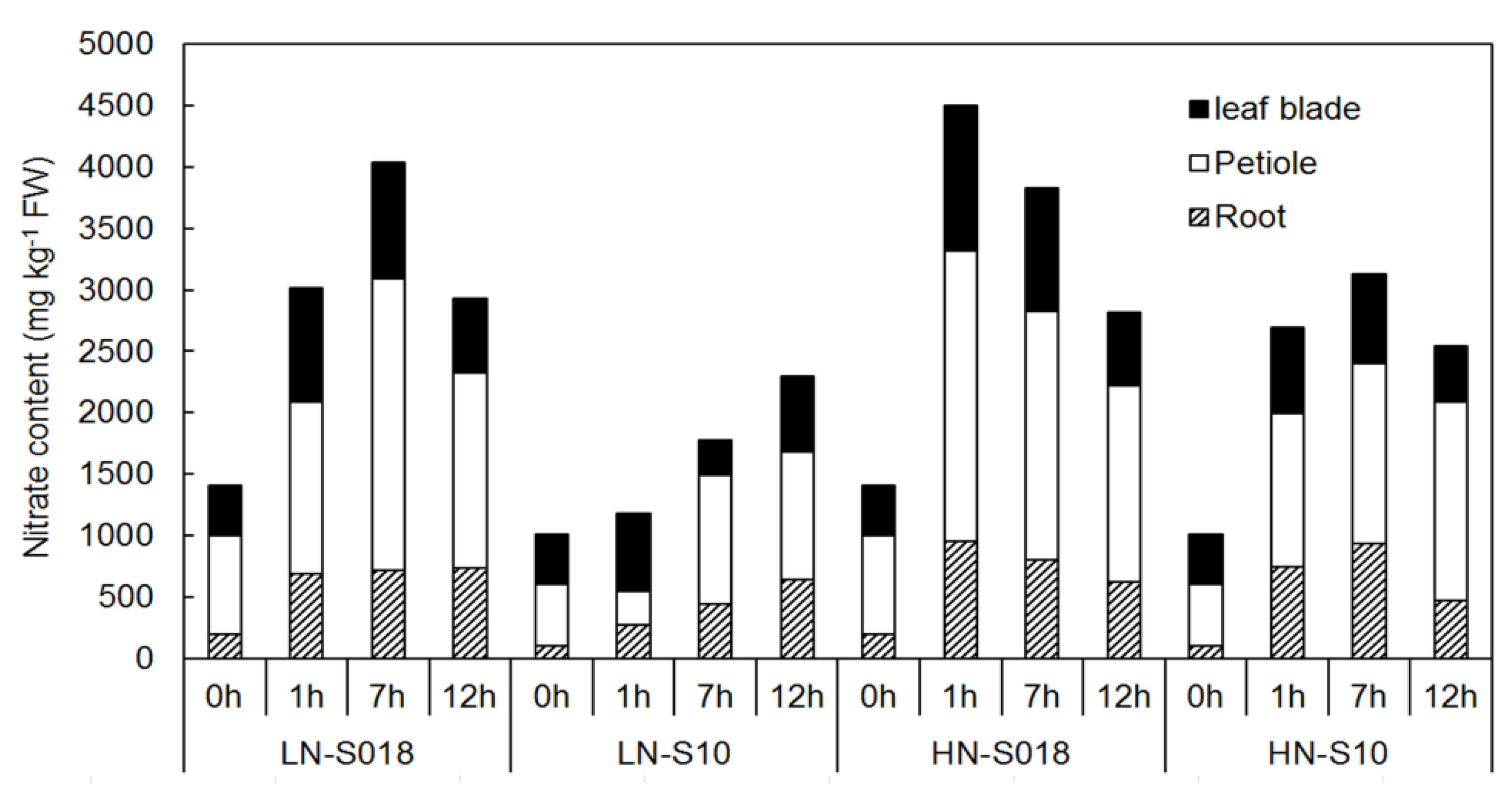
2.1. Fresh Weight and Nitrate Content
2.2. 15NO3−-N Uptake Rate
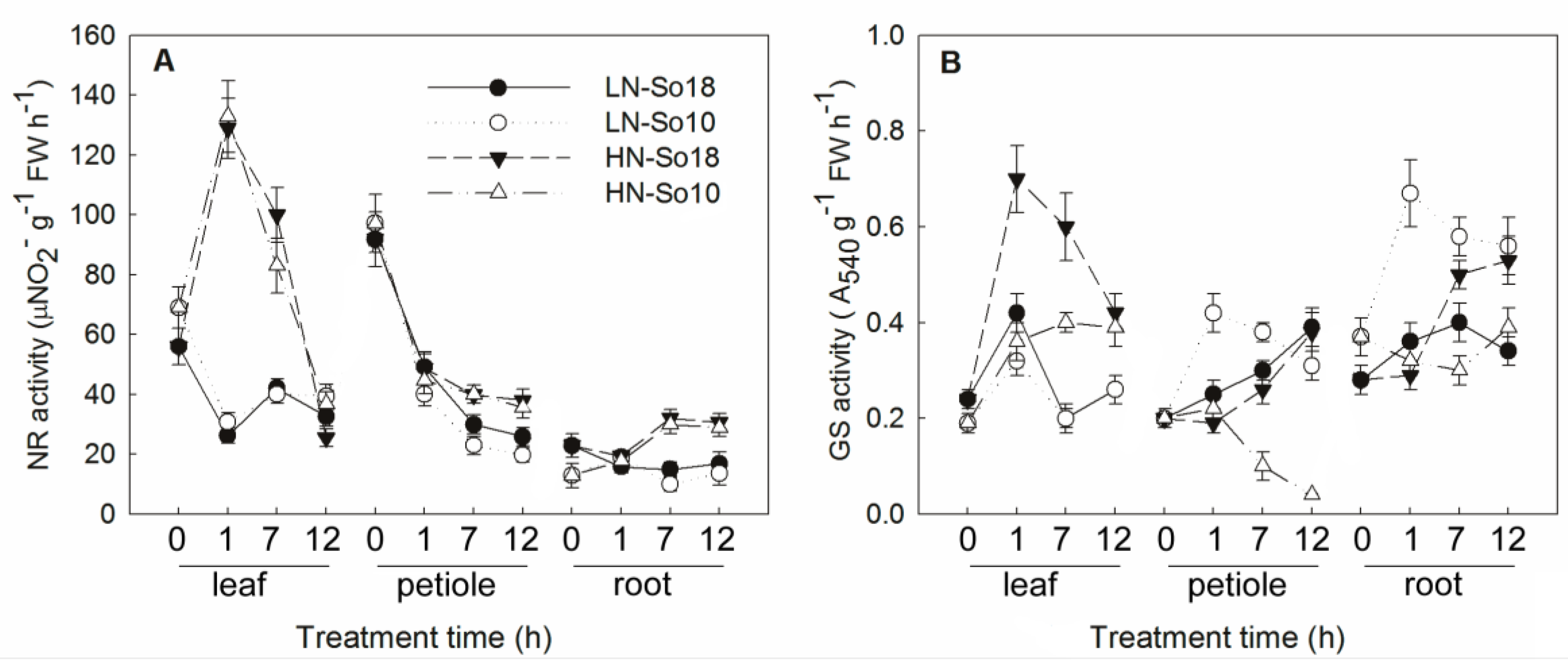
2.3. N Assimilation Related-Enzymes Activities
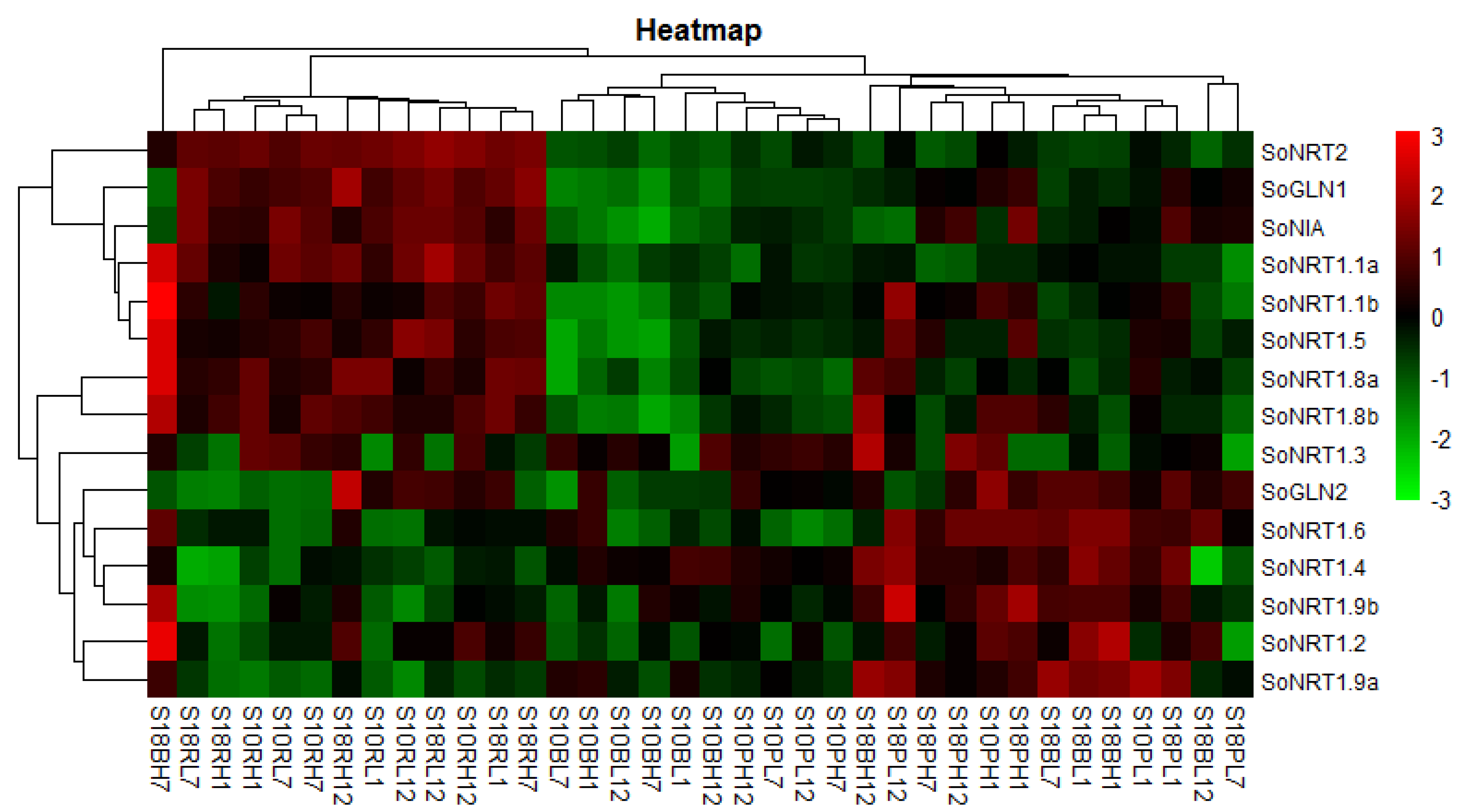
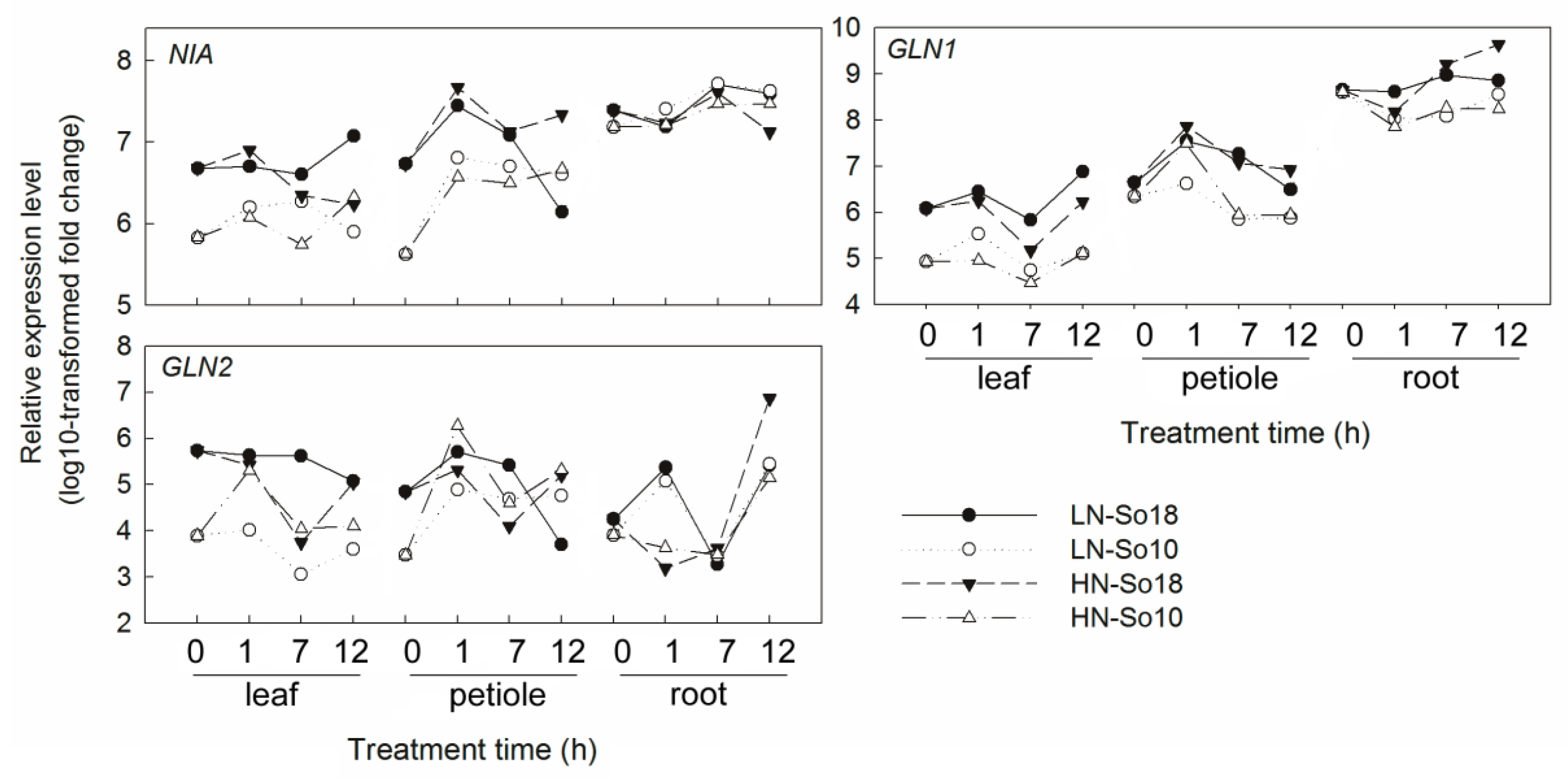
2.4. Expression Analysis of Nitrate Transport and Assimilation Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Sample Preparation
4.2. 15NO3−-N Uptake Rate
4.3. Measurements of Nitrate Content and Enzymes Activity
4.4. Identification of Nitrogen-Responsive Genes and Gene Expression Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walker, R. Nitrates, nitrites and N-nitrosocompounds: A review of the occurrence in food and diet and the toxicological implications. Food Addit. Contam. 1990, 7, 717–768. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, P. Nitrate in vegetables: Toxicity, content, intake and EC regulation. J. Sci. Food Agric. 2006, 86, 10–17. [Google Scholar] [CrossRef]
- Anjana, U.; Iqbal, M. Factors responsible for nitrate accumulation: A review. In Sustainable Agriculture; Lichtfouse, E., Navarrete, M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 533–549. ISBN 978-90-481-2665-1. [Google Scholar]
- Granstedt, R.C.; Huffaker, R.C. Identification of the leaf vacuole as a major nitrate storage pool. Plant Physiol. 1982, 70, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Liu, Q.; Gu, J.; Gong, J.; Guan, C.; Lepo, J.E.; Rong, X.; Song, H.; Zhang, Z. V-ATPase and V-PPase at the tonoplast affect NO3- content in Brassica napus by controlling distribution of NO3− between the cytoplasm and vacuole. J. Plant Growth Regul. 2015, 34, 22–34. [Google Scholar] [CrossRef]
- Anjana, S.U.; Iqbal, M. Nitrate accumulation in plants, factors affecting the and human health implications. A review. Agron. Sustain. Dev. 2007, 27, 45–57. [Google Scholar]
- Mobini, M.; Khoshgoftarmanesh, A.H.; Ghasemi, S. The effect of partial replacement of nitrate with arginine, histidine, and a mixture of amino acids extracted from blood powder on yield and nitrate accumulation in onion bulb. Sci. Hortic. Amsterdam 2014, 176, 232–237. [Google Scholar] [CrossRef]
- Marsic, N.K.; Osvald, J. The influence of different concentration of nitrogen in nutrient solution on plant growth and nitrate accumulation in aeroponically grown lettuce (Lactuca sativa L.). Agrochimica 2002, 46, 56–65. [Google Scholar]
- Onyango, C.M.; Harbinson, J.; Imungi, J.K.; Shibairo, S.S.; van Kooten, O. Influence of organic and mineral fertilization on germination, leaf nitrogen, nitrate accumulation and yield of vegetable amaranth. J. Plant Nutr. 2012, 35, 342–365. [Google Scholar] [CrossRef]
- Inal, A.; Tarakcioglu, C. Effects of nitrogen forms on growth, nitrate accumulation, membrane permeability, and nitrogen use efficiency of hydroponically grown bunch onion under boron deficiency and toxicity. J. Plant Nutr. 2001, 24, 1521–1534. [Google Scholar] [CrossRef]
- Irigoyen, I.; Lamsfus, C.; Aparicio-Tejo, P.; Muro, J. The influence of 3,4-dimethylpyrazole phosphate and dicyandiamide on reducing nitrate accumulation in spinach under Mediterranean conditions. J. Agric. Sci. 2006, 144, 555–562. [Google Scholar] [CrossRef]
- Ma, C.; Feng, S.; Huang, L.; Li, N.; Xu, X.; Zhou, B.; Jiao, K.; Xu, Q.; Li, R.; Herbert, S.J. Exogenous salicylic acid prevents nitrogen dioxide-induced oxidative injury and nitrate accumulation in Brassica campestris L. ssp. chinensis seedlings. J. Hortic. Sci. Biotechnol. 2010, 85, 215–218. [Google Scholar] [CrossRef]
- Strzetelski, P.; Smolen, S.; Rozek, S.; Sady, W. The effect of diverse iodine fertilization on nitrate accumulation and content of selected compounds in radish plants (Raphanus sativus L.). Acta Sci. Pol. Hortic. 2010, 9, 65–73. [Google Scholar]
- Liu, W.K.; Yang, Q.C. Effects of short-term treatment with various light intensities and hydroponic solutions on nitrate concentration of lettuce. Acta Agric. Scand. B-SP 2012, 62, 109–113. [Google Scholar] [CrossRef]
- Agüera, E.; Ruano, D.; Cabello, P.; de la Haba, P. Impact of atmospheric CO2 on growth, photosynthesis and nitrogen metabolism in cucumber (Cucumis sativus L.) plants. J. Plant Physiol. 2006, 163, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.; Charoenprasert, S.; Mitchell, A.E. Effect of organic and conventional cropping systems on ascorbic acid, vitamin C, flavonoids, nitrate, and oxalate in 27 varieties of spinach (Spinacia oleracea L.). J. Agric. Food Chem. 2012, 60, 3144–3150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Tsay, Y.F.; Chiu, C.C.; Tsai, C.B.; Ho, C.H.; Hsu, P.K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Wittgenstein, N.J.; Le, C.H.; Hawkins, B.J.; Ehlting, J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 2014, 14, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The nitrate transporter (NRT) gene family in poplar. PLoS ONE 2013, 8, e72126. [Google Scholar]
- Migocka, M.; Warzybok, A.; Kobus, G. The genomic organization and transcriptional pattern of genes encoding nitrate transporters 1 (NRT1) in cucumber. Plant Soil 2013, 364, 245–260. [Google Scholar] [CrossRef]
- Chiu, C.; Lin, C.; Hsia, A.; Su, R.; Lin, H.; Tsay, Y. Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 2004, 45, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Taochy, C.; Gaillard, I.; Ipotesi, E.; Oomen, R.; Leonhardt, N.; Zimmermann, S.; Peltier, J.-B.; Szponarski, W.; Simonneau, T.; Sentenac, H.; et al. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 2015, 83, 466–479. [Google Scholar] [PubMed] [Green Version]
- Wu, T.; Qin, Z.; Fan, L.; Xue, C.; Zhou, X.; Xin, M.; Du, Y. Involvement of CsNRT1.7 in nitrate recycling during senescence in cucumber. J. Plant Nutr. Soil Sci. 2014, 177, 714–721. [Google Scholar]
- Zhao, S.; Ye, X.; Zhang, Y.; Zheng, J. The contribution of bnnrt1 and bnnrt2 to nitrate accumulation varied according to genotypes in Chinese cabbage. Afr. J. Biotechnol. 2010, 9, 4910–4917. [Google Scholar]
- Quaggiotti, S.; Ruperti, B.; Borsa, P.; Destro, T.; Malagoli, M. Expression of a putative high-affinity NO3− transporter and of an H+-ATPase in relation to whole plant nitrate transport physiology in two maize genotypes differently responsive to low nitrogen availability. J. Exp. Bot. 2003, 54, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- North, K.A.; Ehlting, B.; Koprivova, A.; Rennenberg, H.; Kopriva, S. Natural variation in Arabidopsis adaptation to growth at low nitrogen conditions. Plant Physiol. Biochem. 2009, 47, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Sun, S.; Jia, L.; Chen, W.; Shen, Q. The mechanism of nitrate accumulation in pakchoi [Brassica campestris L. ssp. Chinensis (L.)]. Plant Soil 2006, 282, 291–300. [Google Scholar] [CrossRef]
- Curtis, I.S.; Power, J.B.; de Laat, A.M.M.; Caboche, M.; Davey, M.R. Expression of a chimeric nitrate reductase gene in transgenic lettuce reduces nitrate in leaves. Plant Cell Rep. 1999, 18, 889–896. [Google Scholar] [CrossRef]
- Djennane, S.; Chauvin, J.-E.; Quilleré, I.; Meyer, C.; Chupeau, Y. Introduction and expression of a deregulated tobacco nitrate reductase gene in potato lead to highly reduced nitrate levels in transgenic tubers. Transgen. Res. 2002, 11, 175–184. [Google Scholar] [CrossRef]
- Kaminishi, A.; Kita, N. Seasonal change of nitrate and oxalate concentration in relation to the growth rate of spinach cultivars. Hortscience 2006, 41, 1589–1595. [Google Scholar]
- Gangolli, S.D.; van den Brandt, P.A.; Feron, V.J.; Janzowsky, C.; Koeman, J.H.; Speijers, G.J.; Spiegelhalder, B.; Walker, R.; Wisnok, J.S. Nitrate, nitrite and N-nitroso compounds. Eur. J. Pharmacol. 1994, 292, 1–38. [Google Scholar] [CrossRef]
- Commission Regulation (EC) No 1881/2006 of 19 December 2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs. 2006. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006R1881&rid=1 (accessed on 01.07.2018).
- Menard, C.; Heraud, F.; Volatier, J.L.; Leblanc, J.C. Assessment of dietary exposure of nitrate and nitrite in France. Food Addit. Contam. 2008, 25, 971–988. [Google Scholar] [Green Version]
- Thomson, B.M.; Nokes, C.J.; Cressey, P.J. Intake and risk assessment of nitrate and nitrite from New Zealand foods and drinking water. Food Addit. Contam. 2007, 24, 113–121. [Google Scholar]
- Merino, L.; Darnerud, P.O.; Edberg, U.; Åman, P.; Castillo, M.D.P. Levels of nitrate in Swedish lettuce and spinach over the past 10 years. Food Addit. Contam. 2006, 23, 1283–1289. [Google Scholar]
- Ysart, G.; Clifford, R.; Harrison, N. Monitoring for nitrate in UK-grown lettuce and spinach. Food Addit. Contam. 1999, 16, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Mor, F.; Sahindokuyucu, F.; Erdogan, N. Nitrate and nitrite contents of some vegetables consumed in south province of Turkey. J. Anim. Vet. Adv. 2010, 9, 2013–2016. [Google Scholar]
- Chung, S.Y.; Kim, J.S.; Kim, M.; Hong, M.K.; Lee, J.O.; Kim, C.M.; Song, I.S. Survey of nitrate and nitrite contents of vegetables grown in Korea. Food Addit. Contam. 2003, 20, 621–628. [Google Scholar]
- Zhong, W.; Hu, C.; Wang, M. Nitrate and nitrite in vegetables from north China: Content and intake. Food Addit. Contam. 2002, 19, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Olday, F.C.; Barker, A.V.; Maynard, D.N. A physiological basis for different patterns of nitrate accumulation in two spinach cultivars. J. Am. Soc. Hortic. Sci. 1976, 101, 217–219. [Google Scholar]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, P.K.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.C.; Lin, C.S.; Hsu, P.K.; Lin, S.H.; Tsay, Y.F. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 2009, 21, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Almagro, A.; Lin, S.H.; Tsay, Y.F. Characterization of the Arabidopsis nitrate transporter NRT1. 6 reveals a role of nitrate in early embryo development. Plant Cell 2008, 20, 3289–3299. [Google Scholar] [PubMed]
- Wang, Y.Y.; Tsay, Y.F. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 2011, 23, 1945–1957. [Google Scholar] [PubMed]
- Buchner, P.; Hawkesford, M.J. Complex phylogeny and gene expression patterns of members of the nitrate transporter 1/peptide transporter family (NPF) in wheat. J. Exp. Bot. 2014, 65, 5697–5710. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.R.N.; Imai, A.; Tabata, R.; Shigenobu, S.; Yamaguchi, K.; Yamada, M.; Hasebe, M.; Sawa, S.; Motose, H.; Takahashi, T. Polyamine resistance is increased by mutations in a nitrate transporter gene NRT1.3 (AtNPF6.4) in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 834. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Vidmar, J.J.; Glass, A.D.M. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 2003, 44, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Tischner, R. Nitrate uptake and reduction in higher and lower plants. Plant Cell Environ. 2000, 23, 1005–1024. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Xiang, F.; Zhong, M.; Zhou, L.; Liu, H.; Li, S.; Wang, X. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. UK 2017, 7, e1693. [Google Scholar] [CrossRef] [PubMed]
- Garnica, M.; Houdusse, F.; Zamarreno, A.M.; Garcia-Mina, J.M. Nitrate modifies the assimilation pattern of ammonium and urea in wheat seedlings. J. Sci. Food Agric. 2010, 90, 357–369. [Google Scholar]
- Wu, Y.; Zhang, W.; Xu, L.; Wang, Y.; Zhu, X.; Li, C.; Liu, L. Isolation and molecular characterization of nitrite reductase (RsNiR) gene under nitrate treatments in radish. Sci Hortic-Amsterdam. 2015, 193, 276–285. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, W.; Yang, Y.; Li, Z.; Li, N.; Qi, S.; Crawford, N.M.; Wang, Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. UK 2018, 8, 1487. [Google Scholar] [CrossRef]
- Castaings, L.; Marchive, C.; Meyer, C.; Krapp, A. Nitrogen signalling in Arabidopsis: how to obtain insights into a complex signalling network. J. Exp. Bot. 2011, 62, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Gordonweeks, R.; Shen, Q.; Miller, A.J. Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J. Exp. Bot. 2006, 57, 1333–1340. [Google Scholar] [Green Version]
- Orsel, M.; Eulenburg, K.; Krapp, A.; Daniel-Vedele, F. Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 2004, 219, 714–721. [Google Scholar] [PubMed]
- Tang, Y.; Sun, X.; Hu, C.; Tan, Q.; Zhao, X. Genotypic differences in nitrate uptake, translocation and assimilation of two Chinese cabbage cultivars [Brassica campestris L. ssp. Chinensis (L.)]. Plant Physiol. Biochem. 2013, 70, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S. Principles and Techniques of Plant Pysiological Biochemical Experiment, 3rd ed.; Higher Education Press: Beijing, China, 2015; pp. 129–130. ISBN 978-7-04-039646-1. [Google Scholar]
- Xu, C.; Jiao, C.; Sun, H.; Cai, X.; Wang, X.; Ge, C.; Zheng, Y.; Liu, W.; Sun, X.; Xu, Y. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 2017, 8, e15275. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available |


| Nitrate Concentration Treatment | Gene Symbol | Leaf blade | Petiole | Root | |||
|---|---|---|---|---|---|---|---|
| F Value | P Value | F Value | P Value | F Value | P Value | ||
| 0.5 mmol L−1 | SoNRT1.1a | 65.03 | 0.001 | 70.66 | 0.001 | 1.172 | 0.340 |
| SoNRT1.1b | 165.68 | 0.000 | 17.38 | 0.014 | 32.42 | 0.005 | |
| SoNRT1.2 | 218.12 | 0.000 | 4.42 | 0.103 | 96.52 | 0.001 | |
| SoNRT1.3 | 34.63 | 0.004 | 676.55 | 0.000 | 345.14 | 0.000 | |
| SoNRT1.4 | 1.96 | 0.235 | 35.10 | 0.004 | 1.43 | 0.298 | |
| SoNRT 1.5 | 308.94 | 0.000 | 529.18 | 0.000 | 305.17 | 0.000 | |
| SoNRT1.6 | 1389.78 | 0.000 | 182.38 | 0.000 | 1.87 | 0.243 | |
| SoNRT1.8a | 393.56 | 0.000 | 0.17 | 0.702 | 25.67 | 0.007 | |
| SoNRT1.8b | 291.00 | 0.000 | 175.47 | 0.000 | 38.39 | 0.003 | |
| SoNRT1.9a | 18.23 | 0.013 | 296.94 | 0.000 | 176.73 | 0.000 | |
| SoNRT1.9b | 75.74 | 0.001 | 96.22 | 0.001 | 10.19 | 0.033 | |
| SoNRT2 | 1097.97 | 0.000 | 0.26 | 0.637 | 7.44 | 0.053 | |
| SoNIA | 182.52 | 0.000 | 71.58 | 0.001 | 0.03 | 0.879 | |
| SoGLN1 | 526.29 | 0.000 | 484.55 | 0.000 | 153.67 | 0.000 | |
| SoGLN2 | 1479.40 | 0.000 | 213.74 | 0.000 | 2.07 | 0.223 | |
| 15 mmol·L−1 | SoNRT1.1a | 1684.78 | 0.000 | 40.11 | 0.00 | 0.09 | 0.775 |
| SoNRT1.1b | 915.06 | 0.000 | 0.99 | 0.375 | 1.58 | 0.277 | |
| SoNRT1.2 | 351.99 | 0.000 | 1.33 | 0.313 | 69.87 | 0.001 | |
| SoNRT1.3 | 166.19 | 0.000 | 190.68 | 0.000 | 396.17 | 0.000 | |
| SoNRT1.4 | 191.83 | 0.000 | 39.81 | 0.003 | 7.76 | 0.050 | |
| SoNRT 1.5 | 4024.10 | 0.000 | 255.51 | 0.000 | 357.70 | 0.000 | |
| SoNRT1.6 | 2276.30 | 0.000 | 267.66 | 0.000 | 1.09 | 0.356 | |
| SoNRT1.8a | 3253.03 | 0.000 | 9.88 | 0.035 | 88.67 | 0.001 | |
| SoNRT1.8b | 1447.21 | 0.000 | 7.51 | 0.052 | 0.23 | 0.657 | |
| SoNRT1.9a | 210.99 | 0.000 | 106.84 | 0.000 | 85.43 | 0.001 | |
| SoNRT1.9b | 33.88 | 0.004 | 16.43 | 0.015 | 156.65 | 0.000 | |
| SoNRT2 | 1431.02 | 0.000 | 136.43 | 0.000 | 27.52 | 0.006 | |
| SoNIA | 452.08 | 0.000 | 219.11 | 0.000 | 0.01 | 0.921 | |
| SoGLN1 | 493.49 | 0.000 | 78.36 | 0.001 | 475.91 | 0.000 | |
| SoGLN2 | 556.74 | 0.000 | 0.65 | 0.467 | 3329.69 | 0.000 | |
| Low Nitrate Treatment | High Nitrate Treatment | |||||
|---|---|---|---|---|---|---|
| Leaf Blade | Petiole | Root | Leaf Blade | Petiole | Root | |
| SoNRT1.1a | −0.24 | −0.40 | 0.51 | 0.53 | −0.10 | −0.49 |
| SoNRT1.1b | 0.40 | 0.01 | 0.56 | 0.53 | 0.00 | −0.51 |
| SoNRT1.2 | 0.39 | 0.06 | 0.84 * | 0.78 | 0.08 | −0.69 |
| SoNRT1.3 | 0.00 | −0.15 | 0.33 | −0.75 | −0.93 ** | −0.54 |
| SoNRT1.4 | 0.06 | −0.05 | 0.43 | −0.07 | 0.84 * | −0.51 |
| SoNRT1.5 | 0.29 | 0.00 | 0.82 * | 0.28 | 0.91 * | 0.17 |
| SoNRT1.8a | 0.24 | −0.29 | −0.21 | 0.13 | 0.11 | 0.06 |
| SoNRT1.8b | −0.29 | −0.47 | 0.17 | 0.07 | 0.05 | −0.05 |
| SoNRT1.6 | −0.09 | −0.03 | 0.39 | 0.79 | 0.21 | −0.59 |
| SoNRT1.9a | −0.28 | −0.38 | 0.05 | 0.42 | 0.47 | −0.51 |
| SoNRT1.9b | 0.19 | 0.20 | 0.32 | 0.69 | 0.28 | −0.59 |
| SoNRT2 | −0.11 | −0.26 | 0.61 | 0.56 | −0.32 | −0.62 |
| SoNIA | 0.10 | −0.02 | −0.21 | 0.47 | 0.76 | 0.02 |
| SoGLN1 | 0.51 | 0.18 | 0.36 | 0.26 | 0.37 | −0.17 |
| SoGLN2 | 0.29 | −0.08 | 0.58 | 0.16 | −0.45 | −0.77 |
| Gene | Ortholog Locus | Spinach Source Gene | Spinach Gene Symbol | Position of Predicted Genes | Length of ORF | Length of Protein |
|---|---|---|---|---|---|---|
| AtNRT1.1 | AT1G12110.1 | Spo26413 | SoNRT1.1a | 24047-30641 | 1782 | 593 aa |
| AT1G12110.1 | Spo11551 | SoNRT1.1b | 27689-32901 | 1230 | 409 aa | |
| AtNRT1.2 | AT1G69850.1 | Spo24381 | SoNRT1.2 | 9278773-9283346 | 1767 | 588 aa |
| AtNRT1.3 | AT3G21670.1 | Spo18484 | SoNRT1.3 | 32544817-32547940 | 1737 | 578 aa |
| AtNRT1.4 | AT2G26690.1 | Spo05548 | SoNRT1.4 | 370979-375454 | 1755 | 584 aa |
| AtNRT1.5 | AT4G21680.1 | Spo23405 | SoNRT 1.8 | 43590162-43598583 | 1800 | 599 aa |
| AtNRT1.6/1.71 | AT1G27080.1 | Spo25744 | SoNRT1.6 | 39077918-39086543 | 1875 | 624 aa |
| AtNRT1.8 | AT1G32450.1 | Spo00021 | SoNRT1.5a | 111335-115169 | 1752 | 583 aa |
| AT1G32450.1 | Spo00022 | SoNRT1.5b | 135693-137789 | 1773 | 590 aa | |
| AtNRT1.9 | AT1G18880.1 | Spo20247 | SoNRT1.9a | 9835695-9840007 | 1764 | 587 aa |
| AT1G18880.1 | Spo20248 | SoNRT1.9b | 9830221-9833091 | 1776 | 591 aa | |
| AtNRT2.1 | AT1G08090 | Spo09966 | SoNRT2 | 44925216-44928954 | 1584 | 527 aa |
| AtNIA | AT1G77760 | Spo23607 | SoNIA | 51343436-51348754 | 867 | 288 aa |
| AtGLN1 | AT5G37600 | Spo17102 | SoGLN1 | 9174181-9181171 | 1785 | 594 aa |
| AtGLN2 | AT5G35630 | Spo06798 | SoGLN2 | 19807870-19834770 | 2664 | 887 aa |
| Symbol | Gene ID | Primer Sequence (5′→3′) | |
|---|---|---|---|
| Forward | Reverse | ||
| SoNRT1.1a | Spo26413 | TAAGACTGGCGGTTGG | GACAGAGCATGAAGGAGG |
| SoNRT1.1b | Spo11551 | GTGAGGCATGTGAAAGATTA | CCACCAAGTAGGCAGAGC |
| SoNRT1.2 | Spo24381 | TTGTGGAGGTGCTGGAGA | ACGGCTGATTTAGATGGTGAGA |
| SoNRT1.3 | Spo18484 | TTTGGAGGATTTGTCG | ATGTAAAGAGCAGCGTAT |
| SoNRT1.4 | Spo05548 | GATAGCCGTGAACCTTG | GCGAAAATTGCGACA |
| SoNRT1.5 | Spo23405 | CAATCAGGAAATGGAGGAC | TGTTATGGGACAAAGACG |
| SoNRT1.6 | Spo25744 | CCGTTTCCTCGTTCCAGC | ACCCTTCAGCAAGCCCTA |
| SoNRT1.8a | Spo00021 | CATCTATCGCCGTGTT | GCTTAGACTACTTGCCTCT |
| SoNRT1.8b | Spo00022 | TTTTCGGCGTAGGAGT | TGTGGCGATGTTTGGT |
| SoNRT1. 9a | Spo20247 | GGAGTCCCCTTACGAGTG | TCCTTCTGGGTTTATTTTG |
| SoNRT1.9b | Spo20248 | GTGAGTTGGAGCATCGG | CGTCAGGGGTTATTATCG |
| SoNRT2 | Spo09966 | TTTGCCTTCTCGGTCTC | CGCCGCGAATGTGGATAT |
| SoNIA | Spo23607 | CTTGCCAATTCTGAAGCT | TAGTCCAGGCGTTGATAG |
| SoGLN1 | Spo17102 | GGATATTTCGAGGACAGG | TGGTTTCCAAAGAAGGGT |
| SoGLN2 | Spo06798 | CGGAGAAGGGAATGAAA | CACGGATTGAGCAACCAC |
| 18SrRNA | Spo24625 | CCATAAACGATGCCGACCAG | AGCCTTGCGACCATACTCCC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Cai, X.; Xu, C.; Wang, S.; Dai, S.; Wang, Q. Nitrate Accumulation and Expression Patterns of Genes Involved in Nitrate Transport and Assimilation in Spinach. Molecules 2018, 23, 2231. https://doi.org/10.3390/molecules23092231
Wang X, Cai X, Xu C, Wang S, Dai S, Wang Q. Nitrate Accumulation and Expression Patterns of Genes Involved in Nitrate Transport and Assimilation in Spinach. Molecules. 2018; 23(9):2231. https://doi.org/10.3390/molecules23092231
Chicago/Turabian StyleWang, Xiaoli, Xiaofeng Cai, Chenxi Xu, Shui Wang, Shaojun Dai, and Quanhua Wang. 2018. "Nitrate Accumulation and Expression Patterns of Genes Involved in Nitrate Transport and Assimilation in Spinach" Molecules 23, no. 9: 2231. https://doi.org/10.3390/molecules23092231






