Phosphate-Catalyzed Succinimide Formation from an NGR-Containing Cyclic Peptide: A Novel Mechanism for Deammoniation of the Tetrahedral Intermediate
Abstract
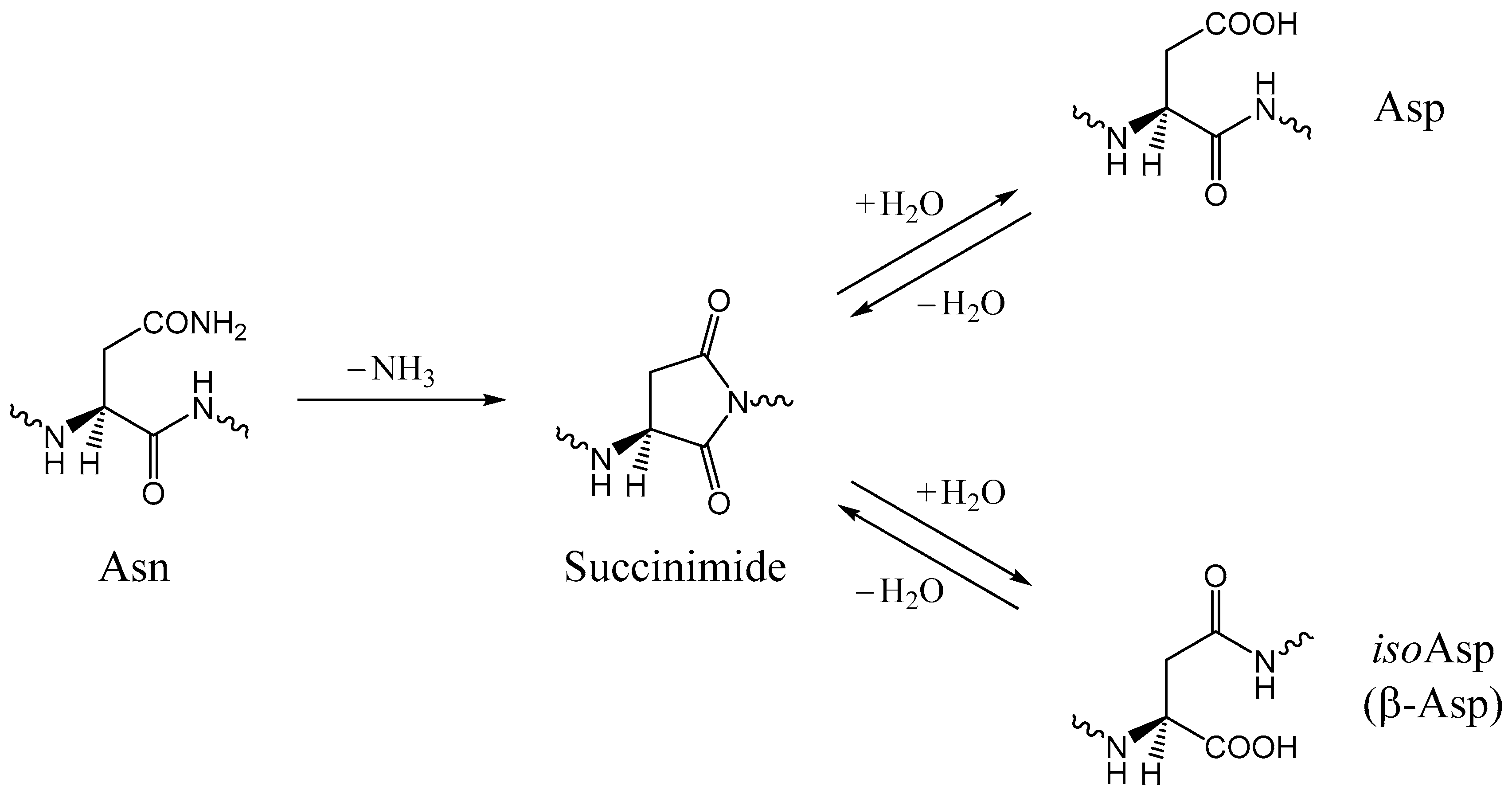
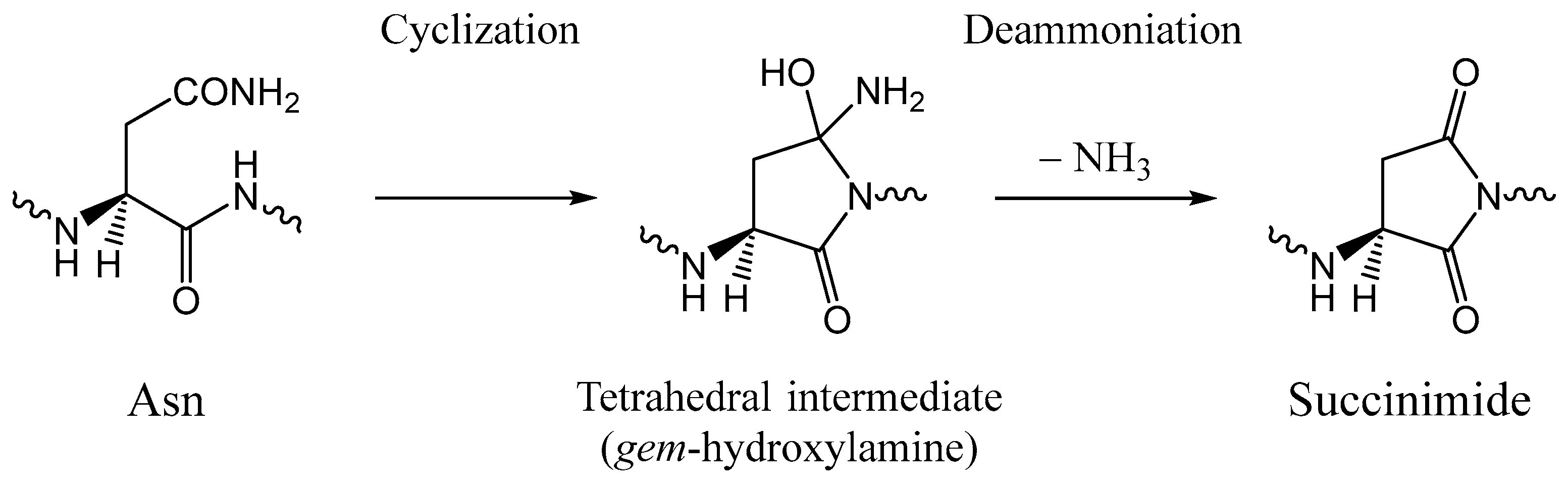
:1. Introduction
2. Results and Discussion
3. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arap, W.; Pasqualini, R.; Ruoslahti, E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 1998, 279, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Ellerby, H.M.; Arap, W.; Ellerby, L.M.; Kain, R.; Andrusiak, R.; Rio, G.D.; Krajewski, S.; Lombardo, C.R.; Rao, R.; Ruoslahti, E.; et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999, 5, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Koivunen, E.; Kain, R.; Lahdenranta, J.; Sakamoto, M.; Stryhn, A.; Ashmun, R.A.; Shapiro, L.H.; Arap, W.; Ruoslahti, E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000, 60, 722–727. [Google Scholar] [PubMed]
- Curnis, F.; Sacchi, A.; Borgna, L.; Magni, F.; Gasparri, A.; Corti, A. Enhancement of tumor necrosis factor α antitumor immunotherapeutic properties by targeted delivery to aminopeptidase N (CD13). Nat. Biotechnol. 2000, 18, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Arrigoni, G.; Sacchi, A.; Fischetti, L.; Arap, W.; Pasqualini, R.; Corti, A. Differential binding of drugs containing the NGR motif to CD13 isoforms in tumor vessels, epithelia, and myeloid cells. Cancer Res. 2002, 62, 867–874. [Google Scholar] [PubMed]
- Pastorino, F.; Brignole, C.; Marimpietri, D.; Cilli, M.; Gambini, C.; Ribatti, D.; Longhi, R.; Allen, T.M.; Corti, A.; Ponzoni, M. Vascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapy. Cancer Res. 2003, 63, 7400–7409. [Google Scholar] [PubMed]
- Sacchi, A.; Gasparri, A.; Curnis, F.; Bellone, M.; Corti, A. Crucial role for interferon γ in the synergism between tumor vasculature-targeted tumor necrosis factor α (NGR-TNF) and doxorubicin. Cancer Res. 2004, 64, 7150–7155. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Gasparri, A.; Sacchi, A.; Cattaneo, A.; Magni, F.; Corti, A. Targeted delivery of IFNγ to tumor vessels uncouples antitumor from counterregulatory mechanisms. Cancer Res. 2005, 65, 2906–2913. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, F.; Brignole, C.; Di Paolo, D.; Nico, B.; Pezzolo, A.; Marimpietri, D.; Pagnan, G.; Piccardi, F.; Cilli, M.; Longhi, R.; et al. Targeting liposomal chemotherapy via both tumor cell-specific and tumor vasculature-specific ligands potentiates therapeutic efficacy. Cancer Res. 2006, 66, 10073–10082. [Google Scholar] [CrossRef] [PubMed]
- Oostendorp, M.; Douma, K.; Hackeng, T.M.; Dirksen, A.; Post, M.J.; van Zandvoort, M.A.M.J.; Backes, W.H. Quantitative molecular magnetic resonance imaging of tumor angiogenesis using cNGR-labeled paramagnetic quantum dots. Cancer Res. 2008, 68, 7676–7683. [Google Scholar] [CrossRef] [PubMed]
- Bieker, R.; Kessler, T.; Schwöppe, C.; Padró, T.; Persigehl, T.; Bremer, C.; Dreischalück, J.; Kolkmeyer, A.; Heindel, W.; Mesters, R.M.; et al. Infarction of tumor vessels by NGR-peptide-directed targeting of tissue factor: Experimental results and first-in-man experience. Blood 2009, 113, 5019–5027. [Google Scholar] [CrossRef] [PubMed]
- Jullienne, B.; Vigant, F.; Muth, E.; Chaligné, R.; Bouquet, C.; Giraudier, S.; Perricaudet, M.; Benihoud, K. Efficient delivery of angiostatin K1-5 into tumors following insertion of an NGR peptide into adenovirus capsid. Gene Ther. 2009, 16, 1405–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Y.; Chen, X.; Wang, J.; Zhang, X.; Zhang, Q. NGR-modified micelles enhance their interaction with CD13-overexpressing tumor and endothelial cells. J. Control. Release 2009, 139, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Gregorc, V.; Citterio, G.; Vitali, G.; Spreafico, A.; Scifo, P.; Borri, A.; Donadoni, G.; Rossoni, G.; Corti, A.; Caligaris-Cappio, F.; et al. Defining the optimal biological dose of NGR-hTNF, a selective vascular targeting agent, in advanced solid tumors. Eur. J. Cancer 2010, 46, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Murase, Y.; Asai, T.; Katanasaka, Y.; Sugiyama, T.; Shimizu, K.; Maeda, N.; Oku, N. A novel DDS strategy, “dual-targeting”, and its application for antineovascular therapy. Cancer Lett. 2010, 287, 165–171. [Google Scholar] [CrossRef] [PubMed]
- van Laarhoven, H.W.M.; Fiedler, W.; Desar, I.M.E.; van Asten, J.J.A.; Marréaud, S.; Lacombe, D.; Govaerts, A.-S.; Bogaerts, J.; Lasch, P.; Timmer-Bonte, J.N.H.; et al. Phase I clinical and magnetic resonance imaging study of the vascular agent NGR-hTNF in patients with advanced cancers (European organization for research and treatment of cancer study 16041). Clin. Cancer Res. 2010, 16, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Gregorc, V.; Zucali, P.A.; Santoro, A.; Ceresoli, G.L.; Citterio, G.; De Pas, T.M.; Zilembo, N.; De Vincenzo, F.; Simonelli, M.; Rossoni, G.; et al. Phase II study of asparagine-glycine-arginine–human tumor necrosis factor α, a selective vascular targeting agent, in previously treated patients with malignant pleural mesothelioma. J. Clin. Oncol. 2010, 28, 2604–2611. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Rimassa, L.; Sobrero, A.F.; Citterio, G.; Sclafani, F.; Carnaghi, C.; Pessino, A.; Caprioni, F.; Andretta, V.; Tronconi, M.C.; et al. Phase II study of NGR-hTNF, a selective vascular targeting agent, in patients with metastatic colorectal cancer after failure of standard therapy. Eur. J. Cancer 2010, 46, 2746–2752. [Google Scholar] [CrossRef] [PubMed]
- Wickström, M.; Larsson, R.; Nygren, P.; Gullbo, J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011, 102, 501–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregorc, V.; De Braud, F.G.; De Pas, T.M.; Scalamogna, R.; Citterio, G.; Milani, A.; Boselli, S.; Catania, C.; Donadoni, G.; Rossoni, G.; et al. Phase I study of NGR-hTNF, a selective vascular targeting agent, in combination with cisplatin in refractory solid tumors. Clin. Cancer Res. 2011, 17, 1964–1972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, B.; Dong, S.; Zhang, Y.; Wu, Y. Anti-tumor efficacy and pre-clinical immunogenicity of IFNα2a-NGR. Regul. Toxicol. Pharmacol. 2011, 60, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Corti, A.; Curnis, F. Tumor vasculature targeting through NGR peptide-based drug delivery systems. Curr. Pharm. Biotechnol. 2011, 12, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Zheng, J.; Rosenblat, J.; Jaffray, D.A.; Allen, C. APN/CD13-targeting as a strategy to alter the tumor accumulation of liposomes. J. Control. Release 2011, 154, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, D.; Scambia, G.; Amadio, G.; di Legge, A.; Pietragalla, A.; De Vincenzo, R.; Masciullo, V.; Di Stefano, M.; Mangili, G.; Citterio, G.; et al. Phase II study of NGR-hTNF in combination with doxorubicin in relapsed ovarian cancer patients. Br. J. Cancer 2012, 107, 37–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corti, A.; Pastorino, F.; Curnis, F.; Arap, W.; Ponzoni, M.; Pasqualini, R. Targeted drug delivery and penetration into solid tumors. Med. Res. Rev. 2012, 32, 1078–1091. [Google Scholar] [CrossRef] [PubMed]
- Soudy, R.; Ahmed, S.; Kaur, K. NGR peptide ligands for targeting CD13/APN identified through peptide array screening resemble fibronectin sequences. ACS Comb. Sci. 2012, 14, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Zucali, P.A.; Simonelli, M.; De Vincenzo, F.; Lorenzi, E.; Perrino, M.; Bertossi, M.; Finotto, R.; Naimo, S.; Balzarini, L.; Bonifacio, C.; et al. Phase I and pharmacodynamic study of high-dose NGR–hTNF in patients with refractory solid tumours. Br. J. Cancer 2013, 108, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Corti, A.; Curnis, F.; Rossoni, G.; Marcucci, F.; Gregorc, V. Peptide-mediated targeting of cytokines to tumor vasculature: The NGR-hTNF example. BioDrugs 2013, 27, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, D.; Pastorino, F.; Zuccari, G.; Caffa, I.; Loi, M.; Marimpietri, D.; Brignole, C.; Perri, P.; Cilli, M.; Nico, B.; et al. Enhanced anti-tumor and anti-angiogenic efficacy of a novel liposomal fenretinide on human neuroblastoma. J. Control. Release 2013, 170, 445–451. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, N.; Caraglia, M.; Grimaldi, A.; Marfella, R.; Servillo, L.; Paolisso, G.; Balestrieri, M.L. Vascular-homing peptides for targeted drug delivery and molecular imaging: Meeting the clinical challenges. Biochim. Biophys. Acta 2014, 1846, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Parmiani, G.; Pilla, L.; Corti, A.; Doglioni, C.; Cimminiello, C.; Bellone, M.; Parolini, D.; Russo, V.; Capocefalo, F.; Maccalli, C. A pilot Phase I study combining peptide-based vaccination and NGR-hTNF vessel targeting therapy in metastatic melanoma. OncoImmunology 2014, 3, e963406. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, Y.; Chen, L.; Lin, Y.-L.; Li, F. A unified mechanism for aminopeptidase N-based tumor cell motility and tumor-homing therapy. J. Biol. Chem. 2014, 289, 34520–34529. [Google Scholar] [CrossRef] [PubMed]
- Zuccari, G.; Milelli, A.; Pastorino, F.; Loi, M.; Petretto, A.; Parise, A.; Marchetti, C.; Minarini, A.; Cilli, M.; Emionite, L.; et al. Tumor vascular targeted liposomal-bortezomib minimizes side effects and increases therapeutic activity in human neuroblastoma. J. Control. Release 2015, 211, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Fiocchi, M.; Sacchi, A.; Gori, A.; Gasparri, A.; Corti, A. NGR-tagged nano-gold: A new CD13-selective carrier for cytokine delivery to tumors. Nano Res. 2016, 9, 1393–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corti, A.; Fiocchi, M.; Curnis, F. Targeting CD13 with Asn-Gly-Arg (NGR) peptide-drug conjugates. In Next-Generation Therapies and Technologies for Immene-Mediated Inflammatory Diseases; Mina-Osorio, P., Ed.; Springer: Cham, Switzerland, 2017; pp. 101–122. ISBN 978-3-319-42252-7. [Google Scholar]
- Roveri, M.; Bernasconi, M.; Leroux, J.-C.; Luciani, P. Peptides for tumor-specific drug targeting: State of the art and beyond. J. Mater. Chem. B 2017, 5, 4348–4364. [Google Scholar] [CrossRef]
- Seidi, K.; Jahanban-Esfahlan, R.; Monhemi, H.; Zare, P.; Minofar, B.; Adli, A.D.F.; Farajzadeh, D.; Behzadi, R.; Abbasi, M.M.; Neubauer, H.A.; et al. NGR (Asn-Gly-Arg)-targetted delivery of coagulase to tumor vasculature arrests cancer cell growth. Oncogene 2018, 37, 3967–3980. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Clarke, S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987, 262, 785–794. [Google Scholar] [PubMed]
- Capasso, S.; Mazzarella, L.; Sica, F.; Zagari, A. Deamidation via cyclic imide in asparaginyl peptides. Pept. Res. 1989, 2, 195–200. [Google Scholar] [PubMed]
- Stephenson, R.C.; Clarke, S. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 1989, 264, 6164–6170. [Google Scholar] [PubMed]
- Patel, K.; Borchardt, R.T. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm. Res. 1990, 7, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Borchardt, R.T. Chemical pathways of peptide degradation. III. Effect of primary sequence on the pathways of deamidation of asparaginyl residues in hexapeptides. Pharm. Res. 1990, 7, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Tyler-Cross, R.; Schirch, V. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J. Biol. Chem. 1991, 266, 22549–22556. [Google Scholar] [PubMed]
- Clarke, S.; Stephenson, R.C.; Lowenson, J.D. Lability of asparagine and aspartic acid residues in proteins and peptides: Spontaneous deamidation and isomerization reactions. In Stability of Protein Pharmaceuticals, Part A: Chemical and Physical Pathways of Protein Degradation; Ahern, T.J., Manning, M.C., Eds.; Plenum Press: New York, NY, USA, 1992; pp. 1–29. ISBN 0-306-44152-7. [Google Scholar]
- Robinson, N.E.; Robinson, Z.W.; Robinson, B.R.; Robinson, A.L.; Robinson, J.A.; Robinson, M.L.; Robinson, A.B. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J. Pept. Res. 2004, 63, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.E.; Robinson, A.B. Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins; Althouse Press: Cave Junction, OR, USA, 2004; ISBN 1-59087-250-0. [Google Scholar]
- Connolly, B.D.; Tran, B.; Moore, J.M.R.; Sharma, V.K.; Kosky, A. Specific catalysis of asparaginyl deamidation by carboxylic acids: Kinetic, thermodynamic, and quantitative structure-property relationship analyses. Mol. Pharm. 2014, 11, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Riggis, D.L.; Gomez, S.V.; Julian, R.R. Sequence and solution effects on the prevalence of d-isomers produced by deamidation. ACS Chem. Biol. 2017, 12, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Manabe, N.; Kirikoshi, R.; Takahashi, O. Glycolic acid-catalyzed deamidation of asparagine residues in degrading PLGA matrices: A computational study. Int. J. Mol. Sci. 2015, 16, 7261–7272. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, O.; Manabe, N.; Kirikoshi, R. A computational study of the mechanism of succinimide formation in the Asn–His sequence: Intramolecular catalysis by the His side chain. Molecules 2016, 21, 327. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, O. Two-water-assisted racemization of the succinimide intermediate formed in proteins: A computational model study. Health 2013, 5, 2018–2021. [Google Scholar] [CrossRef]
- Takahashi, O.; Kirikoshi, R.; Manabe, N. Racemization of the succinimide intermediate formed in proteins and peptides: A computational study of the mechanism catalyzed by dihydrogen phosphate ion. Int. J. Mol. Sci. 2016, 17, 1698. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Longhi, R.; Crippa, L.; Cattaneo, A.; Dondossola, E.; Bachi, A.; Corti, A. Spontaneous formation of l-isoaspartate and gain of function in fibronectin. J. Biol. Chem. 2006, 281, 36466–36476. [Google Scholar] [CrossRef] [PubMed]
- Corti, A.; Curnis, F.; Arap, W.; Pasqualini, R. The neovasculature homing motif NGR: More than meets the eye. Blood 2008, 112, 2628–2635. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Cattaneo, A.; Longhi, R.; Sacchi, A.; Gasparri, A.M.; Pastorino, F.; Di Matteo, P.; Traversari, C.; Bachi, A.; Ponzoni, M.; et al. Critical role of flanking residues in NGR-to-isoDGR transition and CD13/integrin receptor switching. J. Biol. Chem. 2010, 285, 9114–9123. [Google Scholar] [CrossRef] [PubMed]
- Corti, A.; Curnis, F. Isoaspartate-dependent molecular switches for integrin–ligand recognition. J. Cell Sci. 2011, 124, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Zhang, L.; Xie, Y.; Xu, W. NGR-based strategies for targeting delivery of chemotherapeutics to tumor vasculature. Anti-Cancer Agents Med. Chem. 2012, 12, 239–246. [Google Scholar] [CrossRef]
- Enyedi, K.N.; Czajlik, A.; Knapp, K.; Láng, A.; Majer, Z.; Lajkó, E.; Kőhidai, L.; Perczel, A.; Mező, G. Development of cyclic NGR peptides with thioether linkage: Structure and dynamics determining deamidation and bioactivity. J. Med. Chem. 2015, 58, 1806–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enyedi, K.N.; Tóth, S.; Szakács, G.; Mező, G. NGR-peptide–drug conjugates with dual targeting properties. PLoS ONE 2017, 12, e0178632. [Google Scholar] [CrossRef] [PubMed]
- Dutta, B.; Park, J.E.; Kumar, S.; Hao, P.; Gallart-Palau, X.; Serra, A.; Ren, Y.; Sorokin, V.; Lee, C.N.; Ho, H.H.; et al. Monocyte adhesion to atherosclerotic matrix proteins is enhanced by Asn-Gly-Arg deamidation. Sci. Rep. 2017, 7, 5765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripodi, A.A.P.; Tóth, S.; Enyedi, K.N.; Schlosser, G.; Szakács, G.; Mező, G. Development of novel cyclic NGR peptide–daunomycin conjugates with dual targeting property. Beilstein J. Org. Chem. 2018, 14, 911–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastrangeli, R.; D’amici, F.; D’Acunto, C.-W.; Fiumi, S.; Rossi, M.; Terlizzese, M.; Palinsky, W.; Bierau, H. A deamidated interferon-β variant binds to integrin αvβ3. Cytokine 2018, 104, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Spitaleri, A.; Mari, S.; Curnis, F.; Traversari, C.; Longhi, R.; Bordignon, C.; Corti, A.; Rizzardi, G.-P.; Musco, G. Structural basis for the interaction of isoDGR with the RGD-binding site of αvβ3 integrin. J. Biol. Chem. 2008, 283, 19757–19768. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Sacchi, A.; Gasparri, A.; Longhi, R.; Bachi, A.; Doglioni, C.; Bordignon, C.; Traversari, C.; Rizzardi, G.-P.; Corti, A. Isoaspartate-glycine-arginine: A new tumor vasculature-targeting motif. Cancer Res. 2008, 68, 7073–7082. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cheng, Q.; Jin, X.; Ji, J.-L.; Guo, S.; Zheng, S.; Wang, X.; Cao, H.; Gao, S.; Liang, X.-J.; et al. Systemic and tumor-targeted delivery of siRNA by cyclic NGR and isoDGR motif-containing peptides. Biomater. Sci. 2016, 4, 494–510. [Google Scholar] [CrossRef] [PubMed]
- Kirikoshi, R.; Manabe, N.; Takahashi, O. Phosphate-catalyzed succinimide formation from Asp residues: A computational study of the mechanism. Int. J. Mol. Sci. 2018, 19, 637. [Google Scholar] [CrossRef] [PubMed]
- Kirikoshi, R.; Manabe, N.; Takahashi, O. Succinimide formation from an NGR-containing cyclic peptide: Computational evidence for catalytic roles of phosphate buffer and the arginine side chain. Int. J. Mol. Sci. 2017, 18, 429. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.D.; Neilson, G.W.; Enderby, J.E. Ions in water: Characterizing the forces that control chemical processes and biological structure. Biophys. Chem. 2007, 128, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Marenich, A.V.; Olson, R.M.; Kelly, C.P.; Cramer, C.J.; Truhlar, D.G. Self-consistent reaction field model for aqueous and nonaqueous solutions based on accurate polarized partial charges. J. Chem. Theory Comput. 2007, 3, 2011–2033. [Google Scholar] [CrossRef] [PubMed]
- Cramer, C.J.; Truhlar, D.G. A universal approach to solvation modeling. Acc. Chem. Res. 2008, 41, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, O.; Kirikoshi, R.; Manabe, N. Racemization of serine residues catalyzed by dihydrogen phosphate ion: A computational study. Catalysts 2017, 7, 363. [Google Scholar] [CrossRef]
- Spartan ’14, version 1.1.4; Wavefunction, Inc.: Irvine, CA, USA, 2014.
Sample Availability: Samples of the compounds are not available from the authors. |

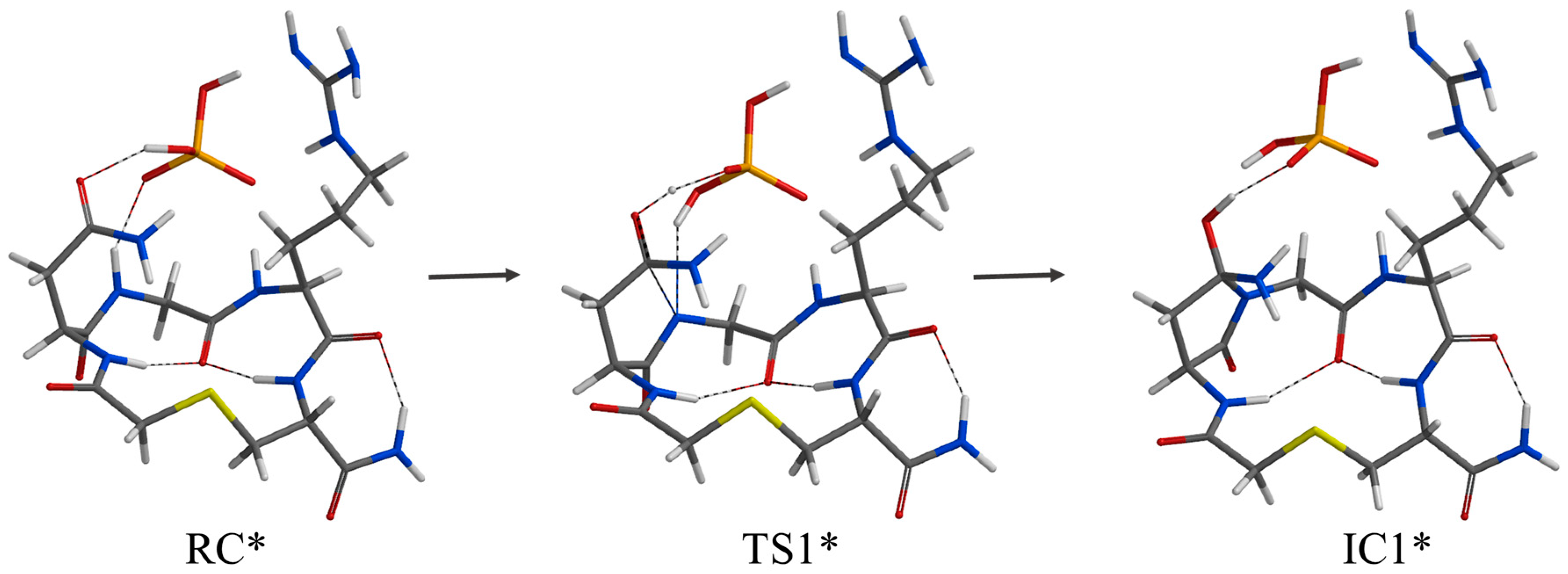
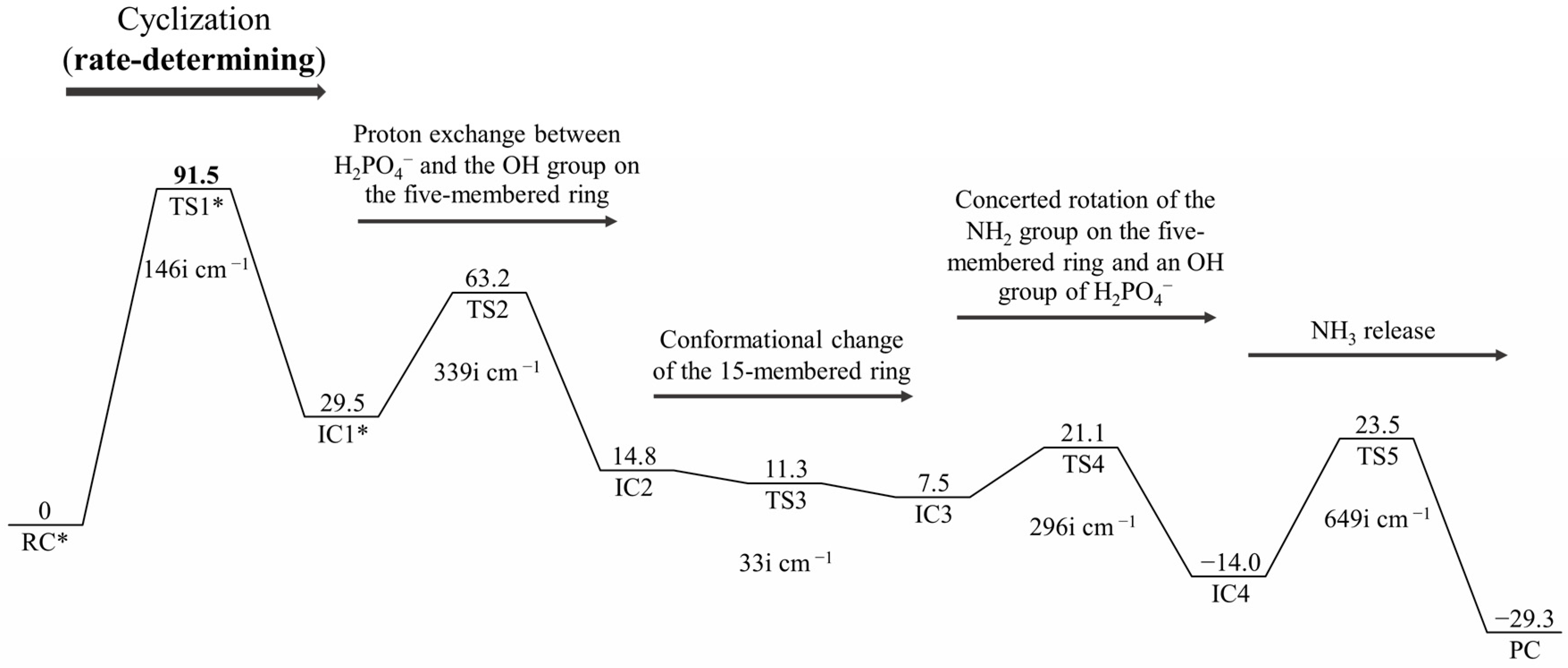
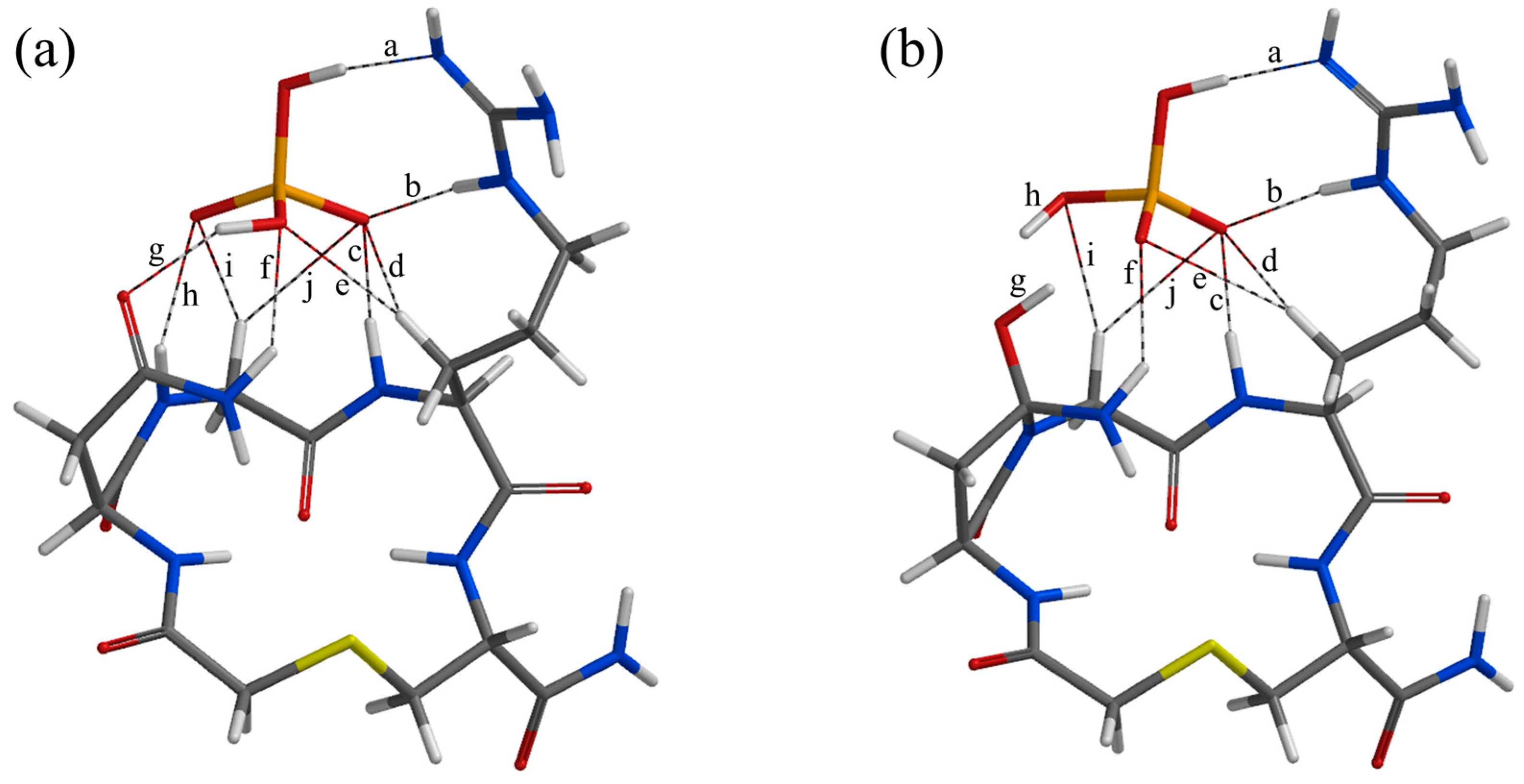

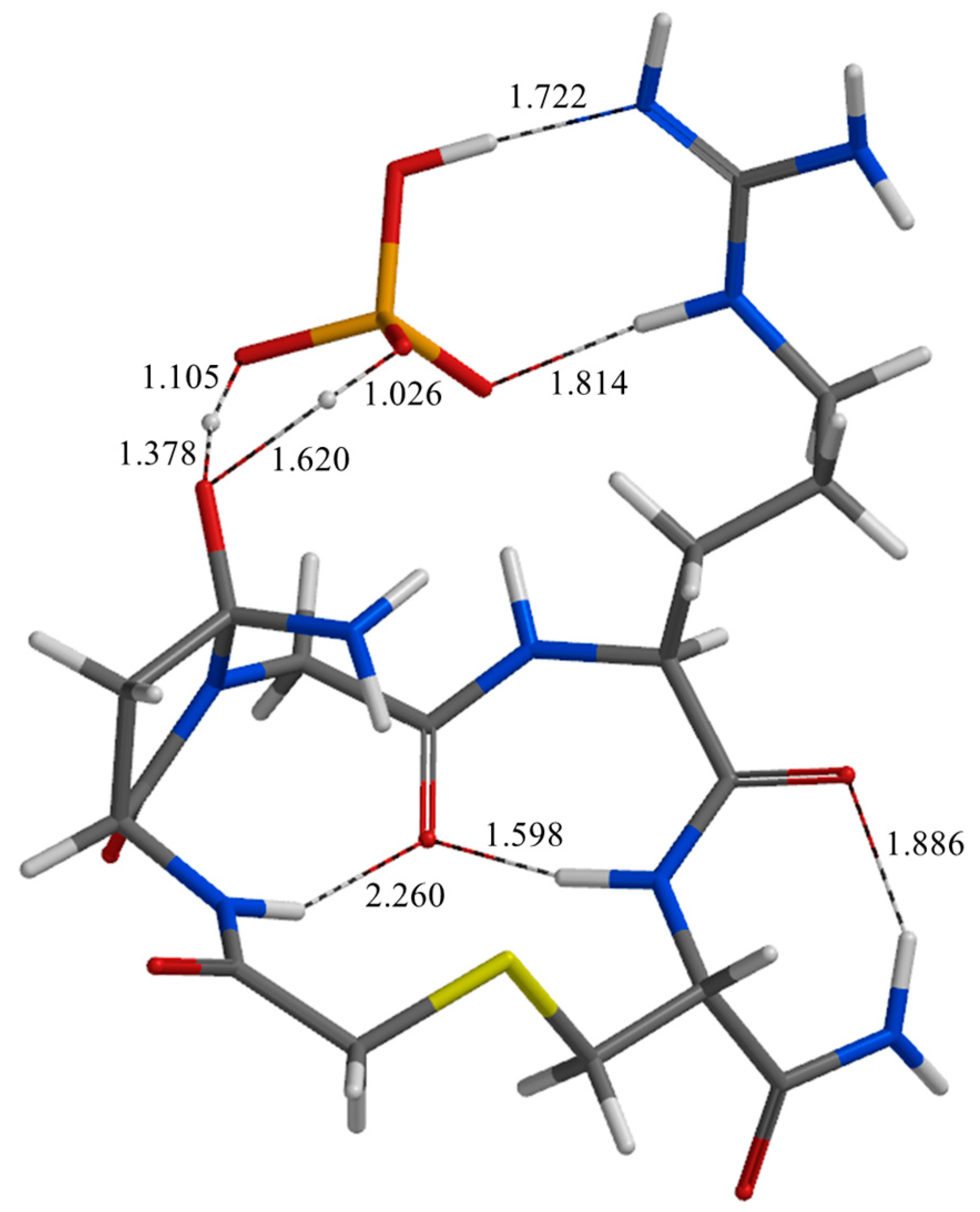
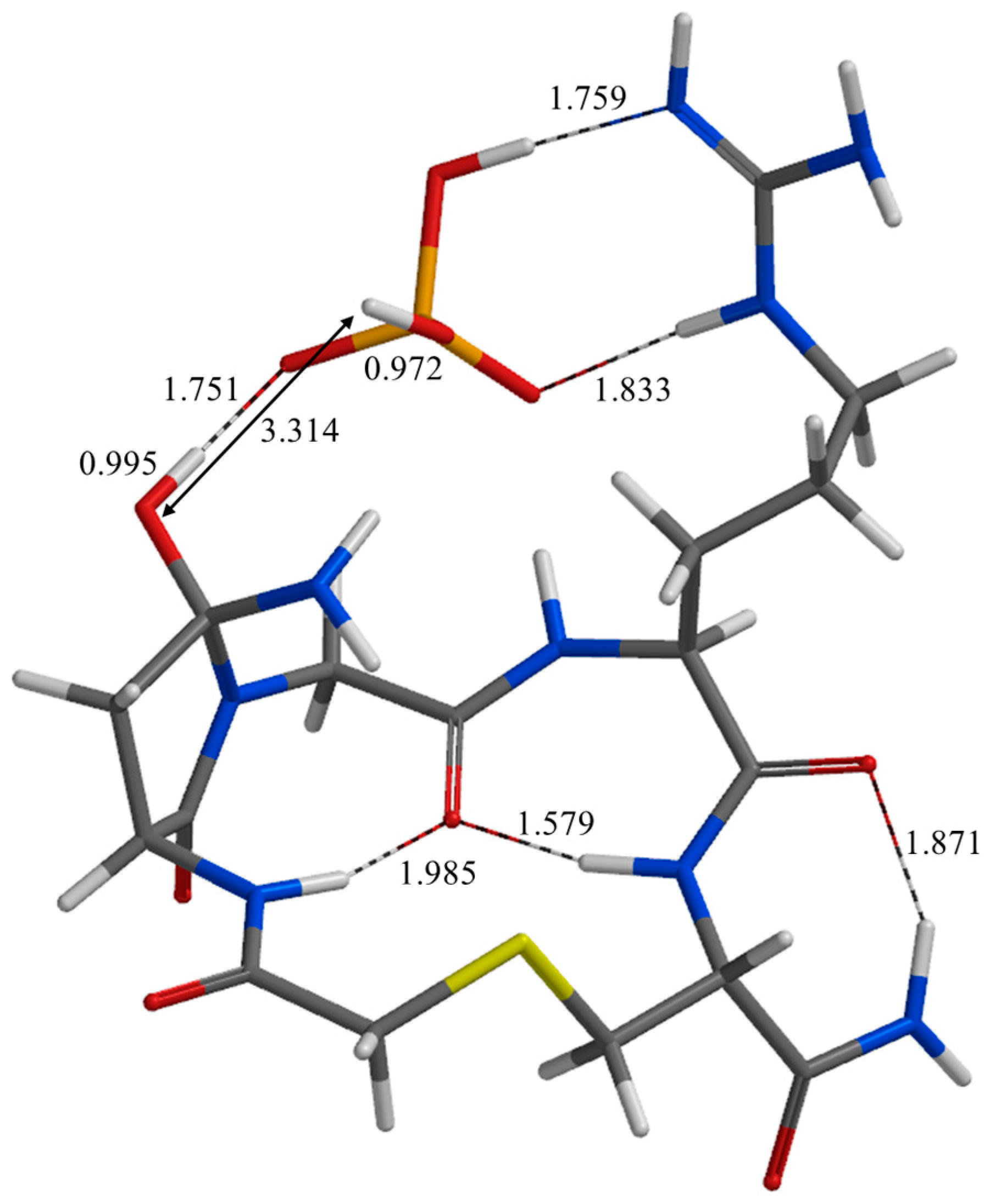
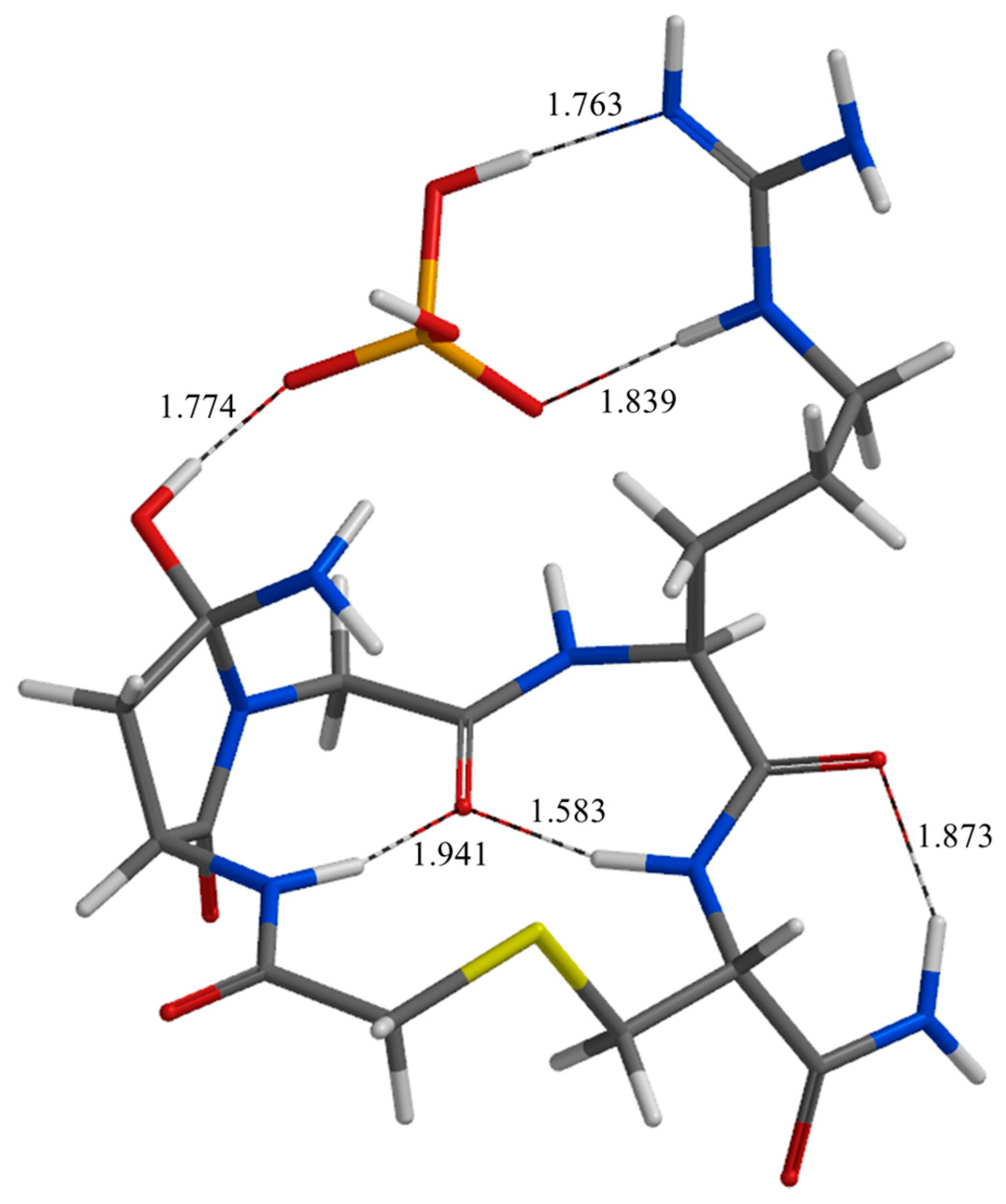
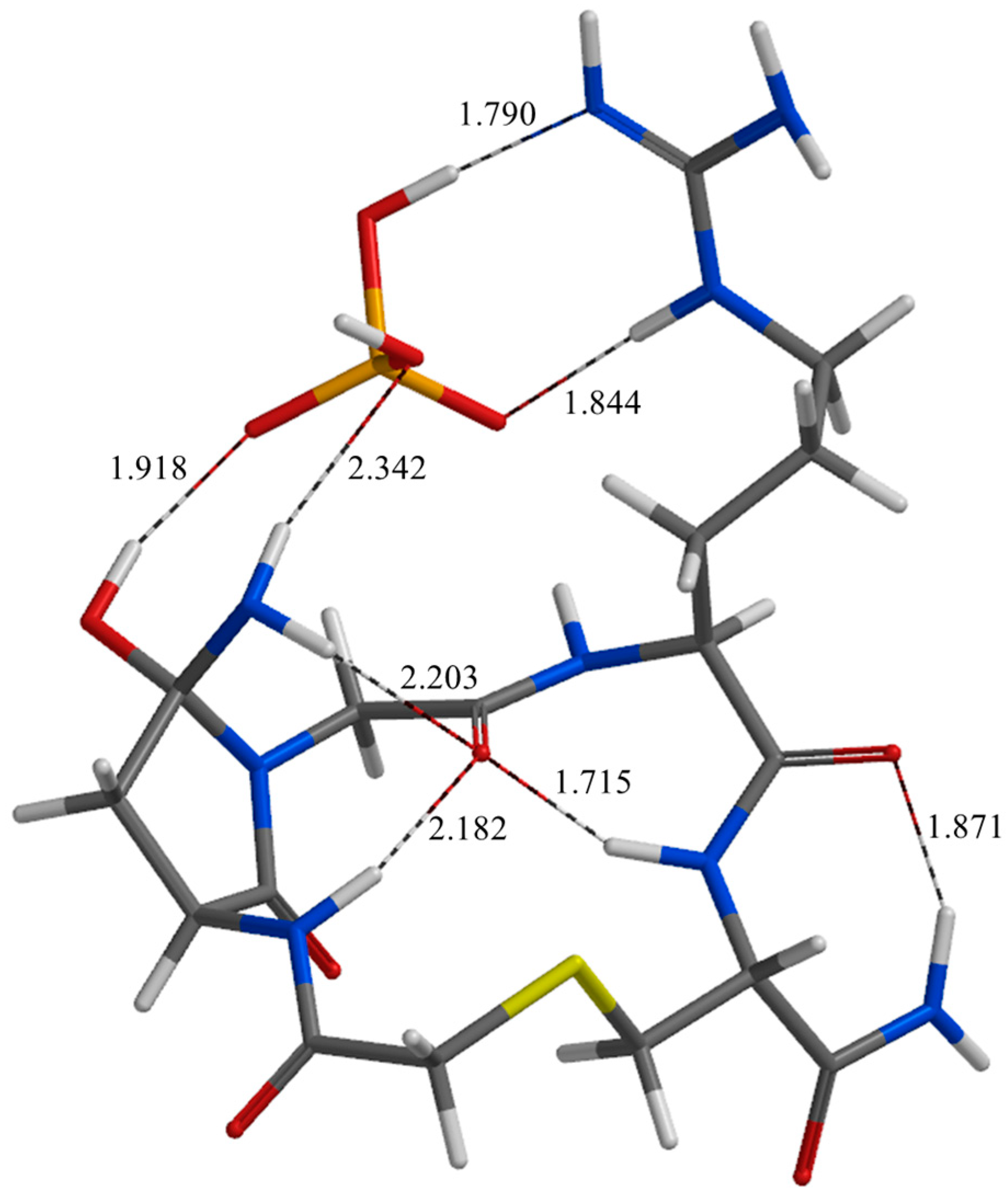
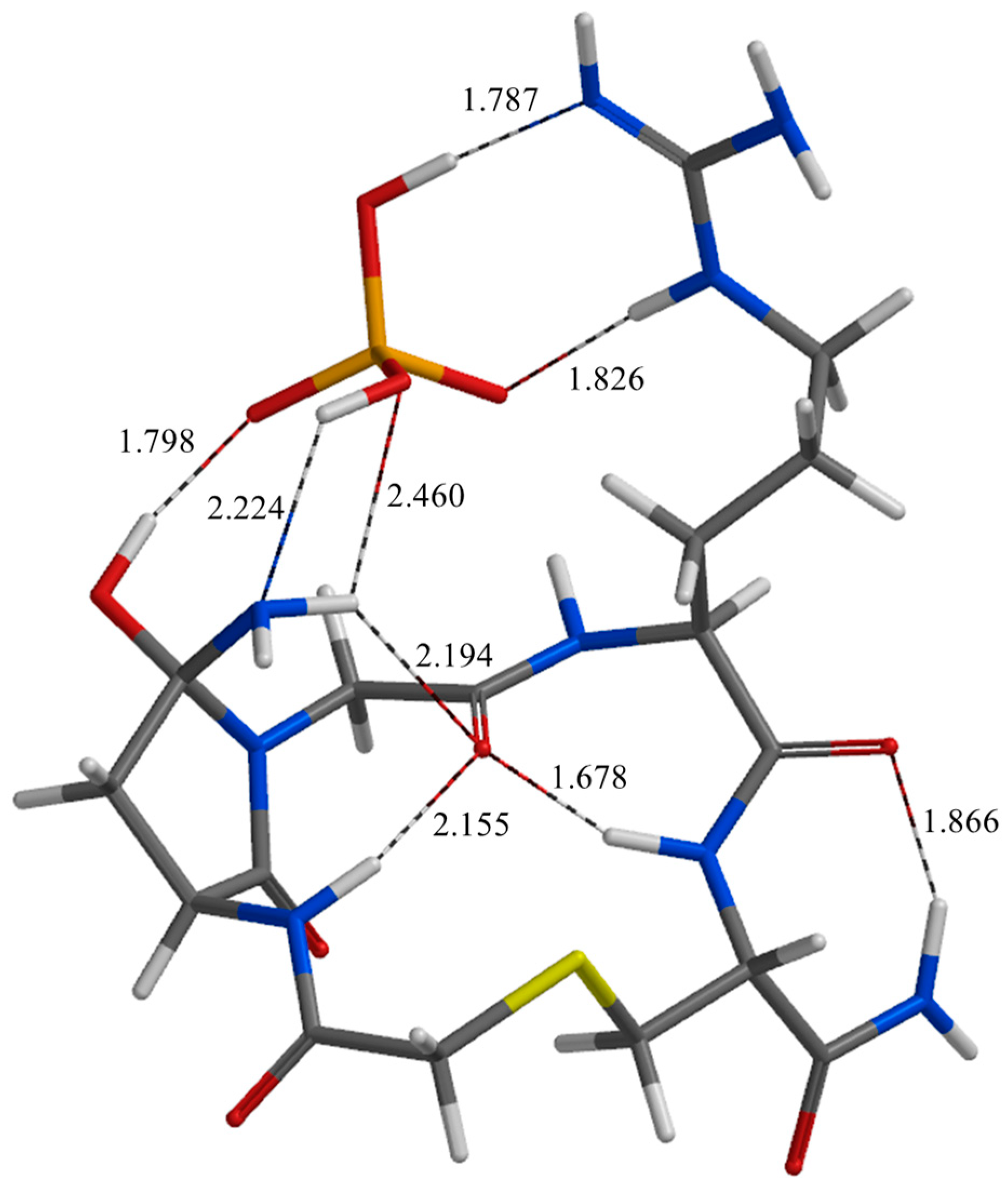
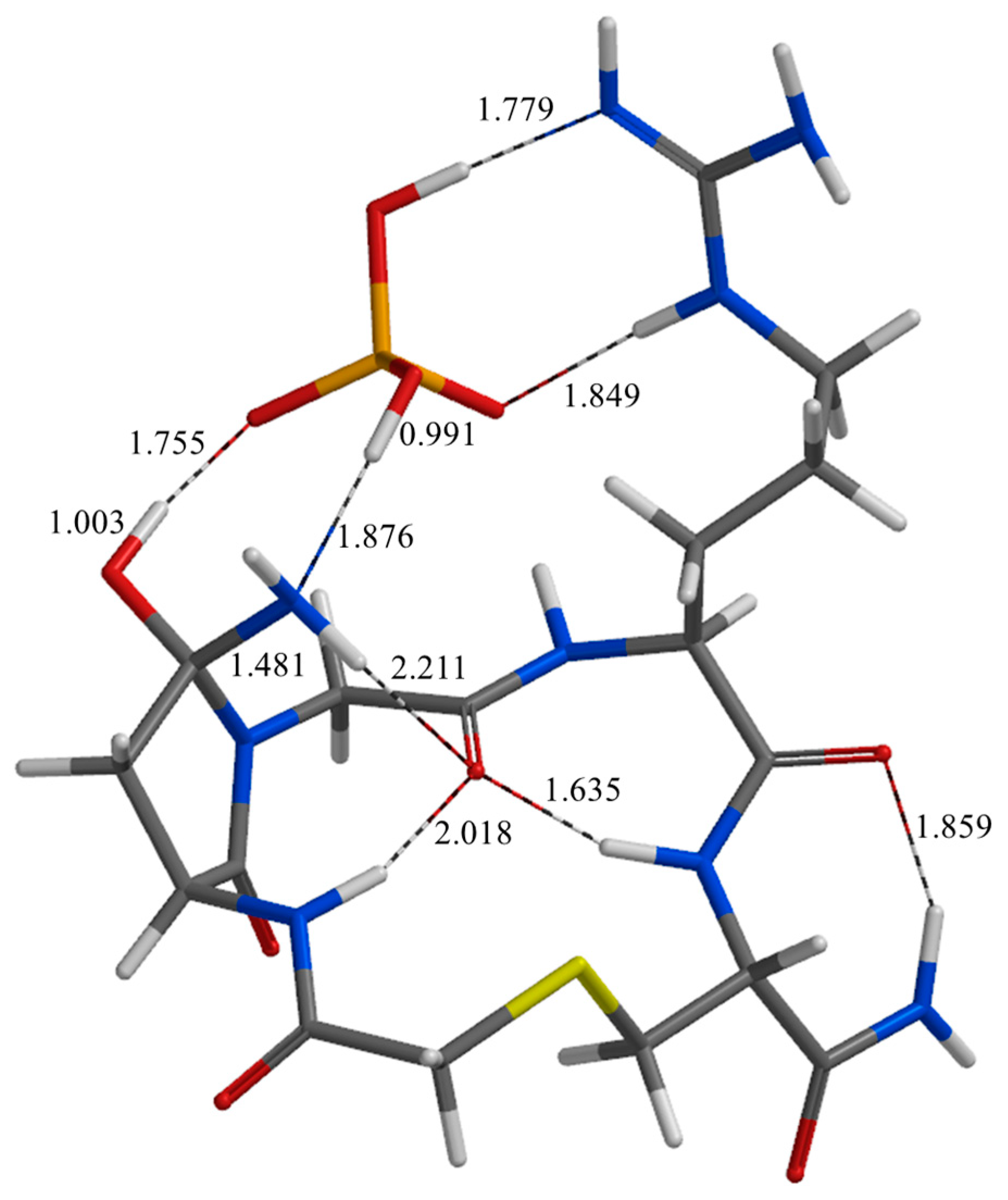
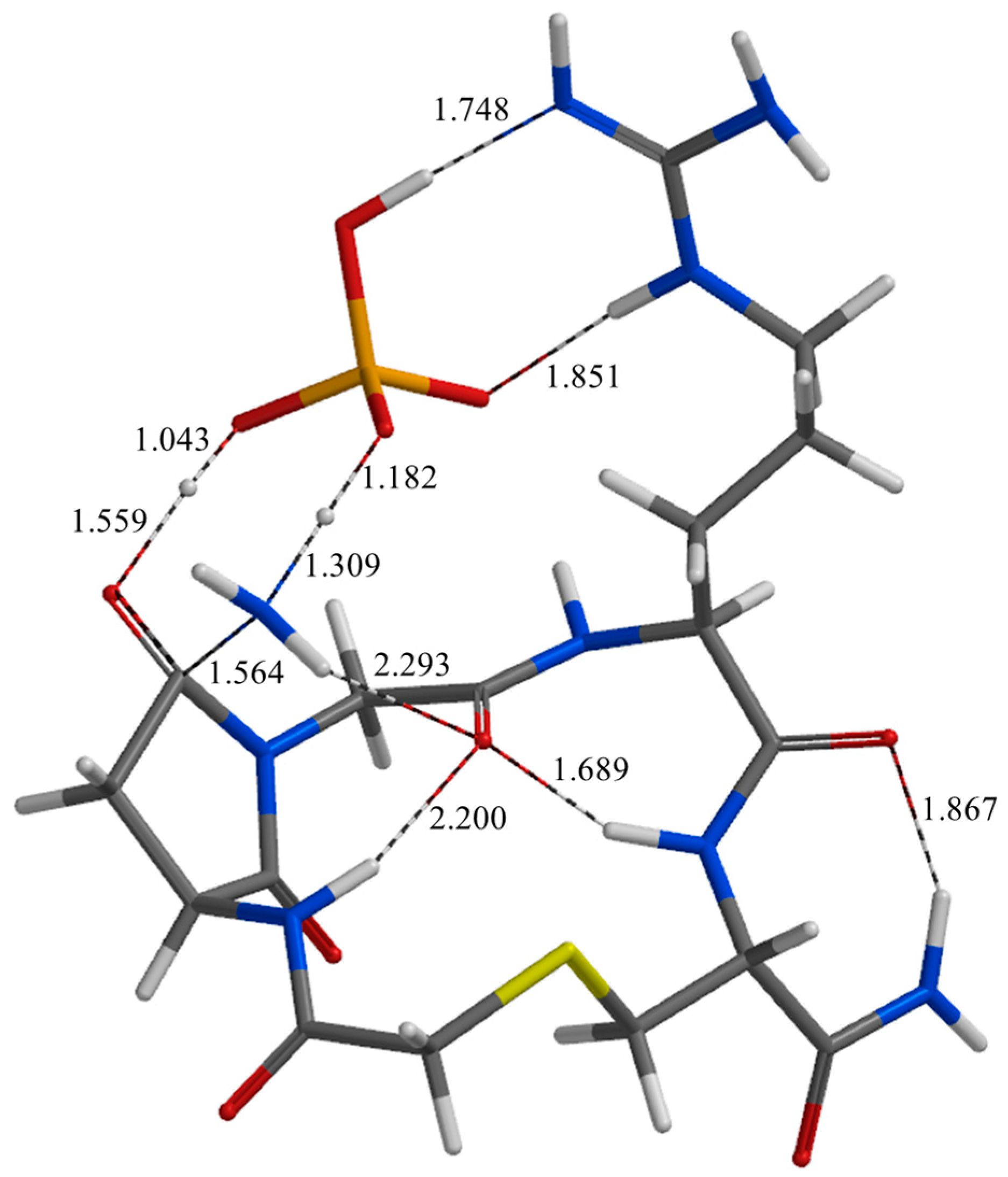
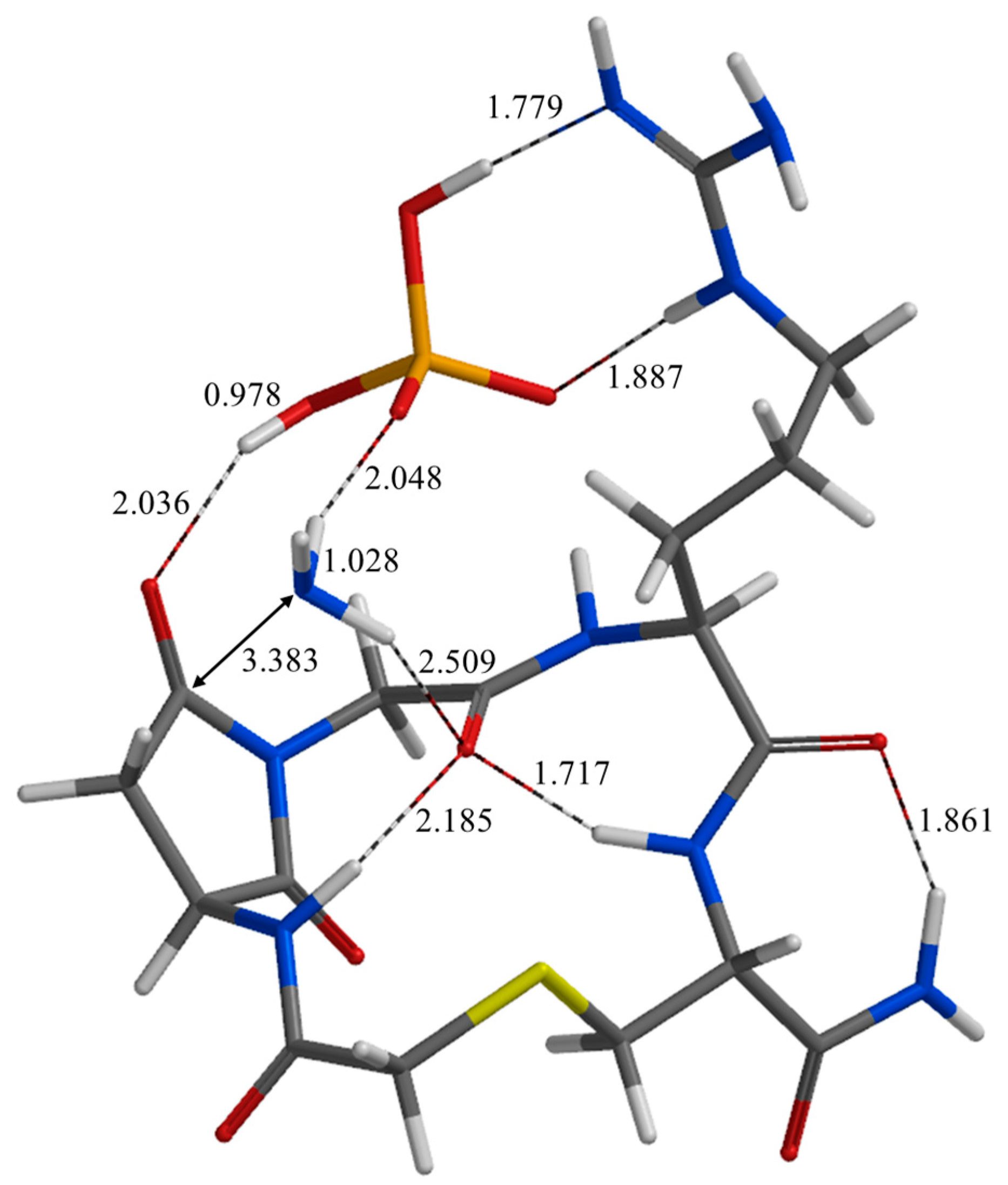
| Geometry | a | b | c | d | e | f | g | h | i | j |
|---|---|---|---|---|---|---|---|---|---|---|
| RC* | 1.824 | 1.789 | 1.713 | 2.361 | 2.421 | 2.196 | 1.893 | 2.235 | 2.306 | 2.911 |
| TS1* | 1.745 | 1.828 | 1.805 | 2.482 | 2.584 | 2.135 | 1.070 | 0.978 | 2.531 | 2.641 |
| IC1* | 1.747 | 1.840 | 1.750 | 2.509 | 2.738 | 2.104 | 1.001 | 0.977 | 2.355 | 2.663 |
| TS2 | 1.722 | 1.814 | 1.794 | 2.456 | 2.752 | 2.472 | 1.620 | 1.105 | 2.359 | 2.586 |
| IC2 | 1.759 | 1.833 | 1.759 | 2.508 | 2.423 | 2.151 | 3.314 | 1.751 | 2.274 | 2.822 |
| TS3 | 1.763 | 1.839 | 1.778 | 2.543 | 2.418 | 2.163 | 3.528 | 1.774 | 2.236 | 2.804 |
| IC3 | 1.790 | 1.844 | 1.932 | 2.751 | 2.475 | 2.342 | 4.321 | 1.918 | 2.161 | 2.726 |
| TS4 | 1.787 | 1.826 | 1.904 | 2.688 | 2.468 | 2.460 | 3.345 | 1.798 | 2.096 | 2.811 |
| IC4 | 1.779 | 1.849 | 1.801 | 2.714 | 2.435 | 3.360 | 3.139 | 1.755 | 2.064 | 2.759 |
| TS5 | 1.748 | 1.851 | 2.030 | 2.915 | 2.444 | 2.968 | 2.700 | 1.043 | 2.232 | 2.737 |
| PC | 1.779 | 1.887 | 1.762 | 2.812 | 2.297 | 3.236 | 3.041 | 0.978 | 2.185 | 3.036 |
| Geometry | θ1 | θ2 | θ3 | θ4 | θ5 | θ6 | θ7 | θ8 | θ9 | θ10 | θ11 | θ12 | θ13 | θ14 | θ15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RC* | −146 | −91 | 139 | −78 | −121 | 165 | 40 | −33 | −157 | 155 | −63 | 159 | −131 | 44 | 167 |
| TS1* | −165 | −89 | 145 | −91 | −120 | 168 | 20 | −18 | −165 | 160 | −64 | 158 | −132 | 60 | 174 |
| IC1* | −172 | −91 | 149 | −85 | −133 | 169 | 15 | −14 | −165 | 159 | −63 | 165 | −127 | 53 | −179 |
| TS2 | −173 | −91 | 151 | −89 | −126 | 170 | 6 | −9 | −167 | 159 | −64 | 165 | −130 | 58 | −178 |
| IC2 | −152 | −92 | 148 | −78 | −142 | 167 | 31 | −23 | −163 | 160 | −65 | 168 | −116 | 31 | 175 |
| TS3 | −142 | −95 | 148 | −76 | −151 | 165 | 43 | −25 | −162 | 159 | −68 | 169 | −111 | 20 | 173 |
| IC3 | −114 | −105 | 153 | −68 | 177 | 174 | 66 | −28 | −169 | 161 | −75 | 175 | −78 | −23 | 168 |
| TS4 | −115 | −100 | 155 | −70 | −179 | 174 | 66 | −32 | −166 | 161 | −74 | 175 | −80 | −23 | 166 |
| IC4 | −121 | −99 | 151 | −74 | −171 | 172 | 60 | −29 | −165 | 161 | −73 | 173 | −91 | −8 | 168 |
| TS5 | −116 | −101 | 157 | −74 | 180 | 178 | 64 | −28 | −167 | 161 | −76 | 176 | −81 | −19 | 164 |
| PC | −107 | −113 | 147 | −63 | −180 | 180 | 65 | −34 | −169 | 162 | −73 | 173 | −75 | −29 | 170 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirikoshi, R.; Manabe, N.; Takahashi, O. Phosphate-Catalyzed Succinimide Formation from an NGR-Containing Cyclic Peptide: A Novel Mechanism for Deammoniation of the Tetrahedral Intermediate. Molecules 2018, 23, 2217. https://doi.org/10.3390/molecules23092217
Kirikoshi R, Manabe N, Takahashi O. Phosphate-Catalyzed Succinimide Formation from an NGR-Containing Cyclic Peptide: A Novel Mechanism for Deammoniation of the Tetrahedral Intermediate. Molecules. 2018; 23(9):2217. https://doi.org/10.3390/molecules23092217
Chicago/Turabian StyleKirikoshi, Ryota, Noriyoshi Manabe, and Ohgi Takahashi. 2018. "Phosphate-Catalyzed Succinimide Formation from an NGR-Containing Cyclic Peptide: A Novel Mechanism for Deammoniation of the Tetrahedral Intermediate" Molecules 23, no. 9: 2217. https://doi.org/10.3390/molecules23092217






