Comparative Studies of the Dynamics Effects of BAY60-2770 and BAY58-2667 Binding with Human and Bacterial H-NOX Domains
Abstract
:1. Introduction
2. Results and Discussion
2.1. Comparative Modeling
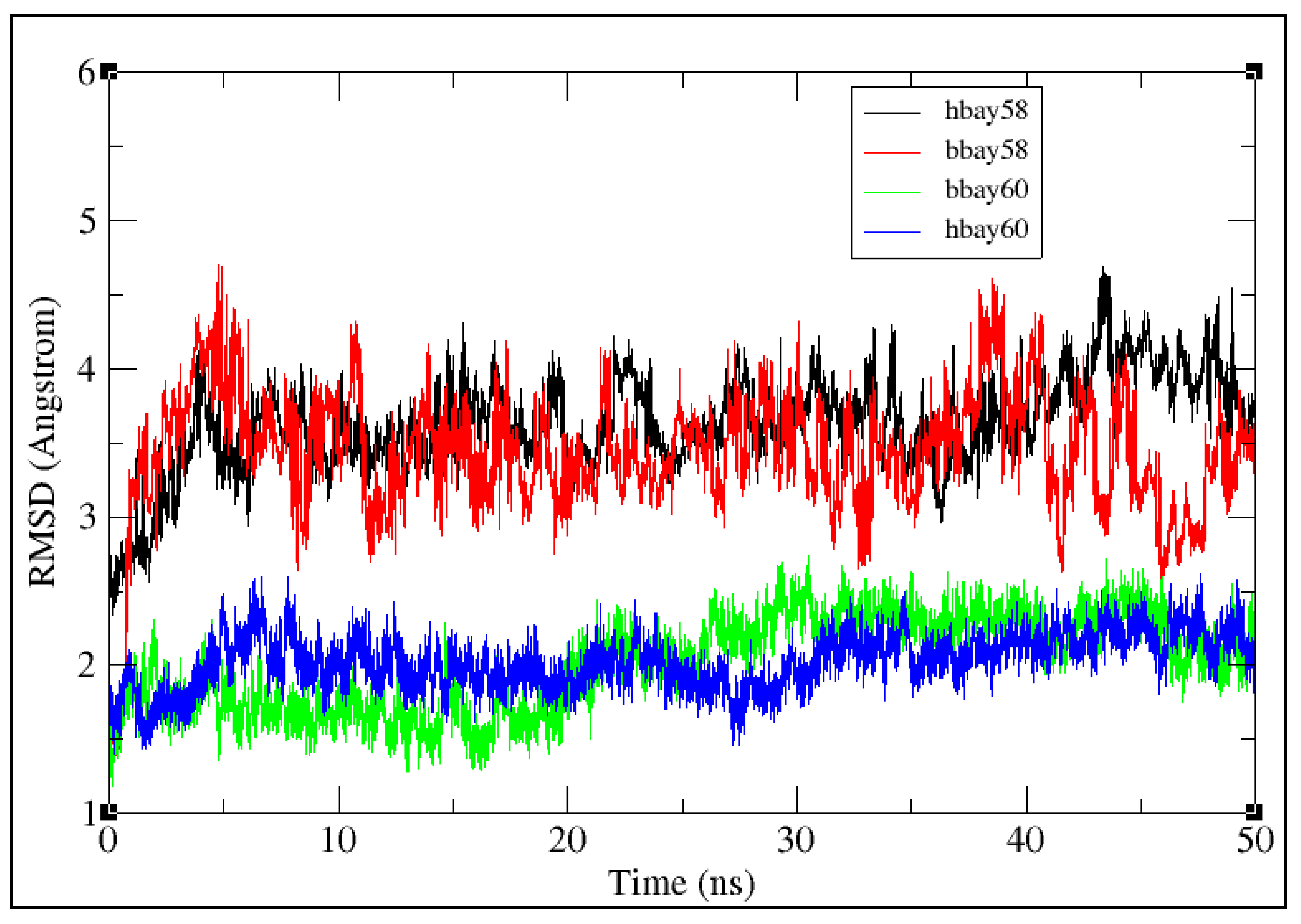
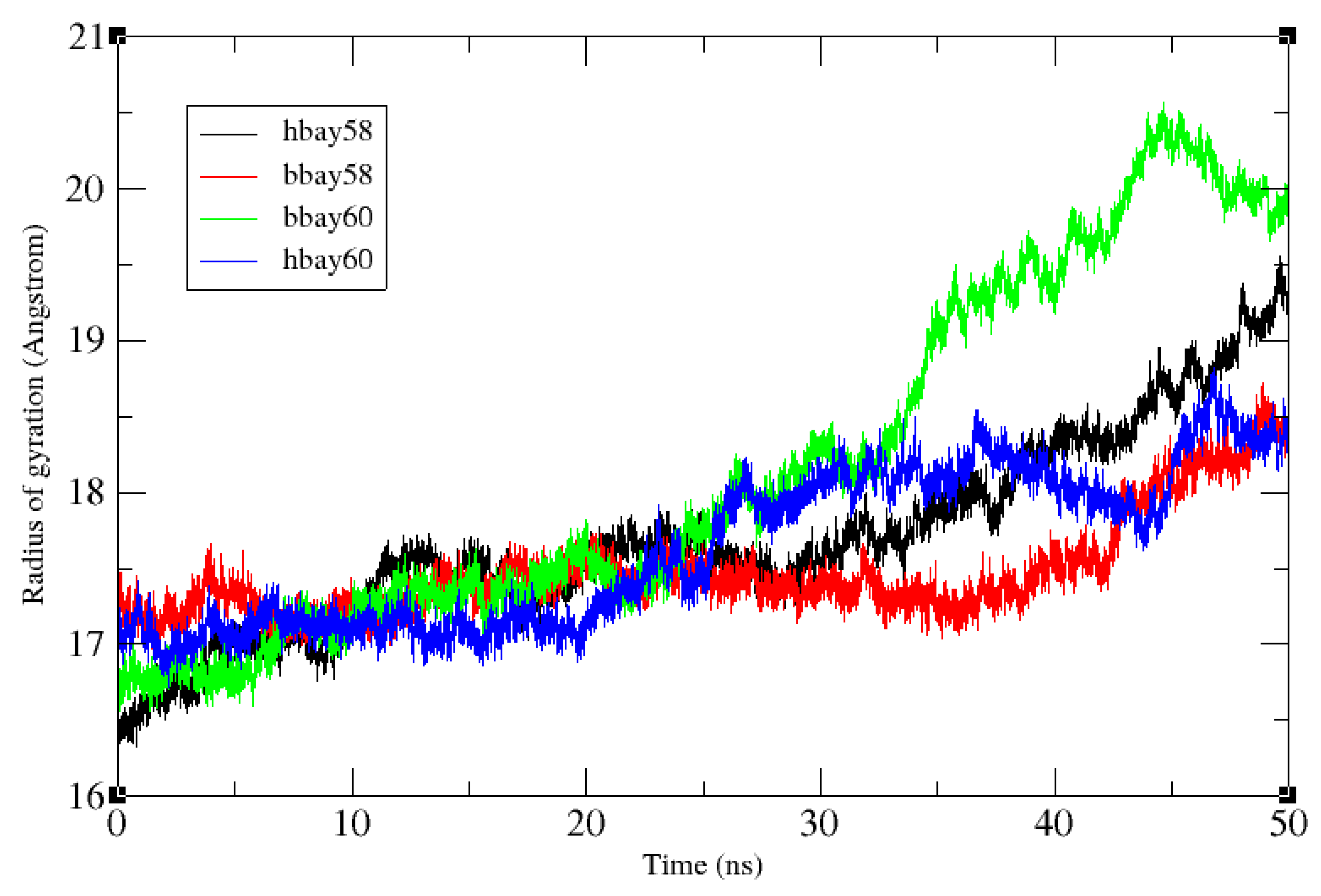
2.2. Molecular Dynamics Simulation
2.3. Hydrogen Bond Analysis
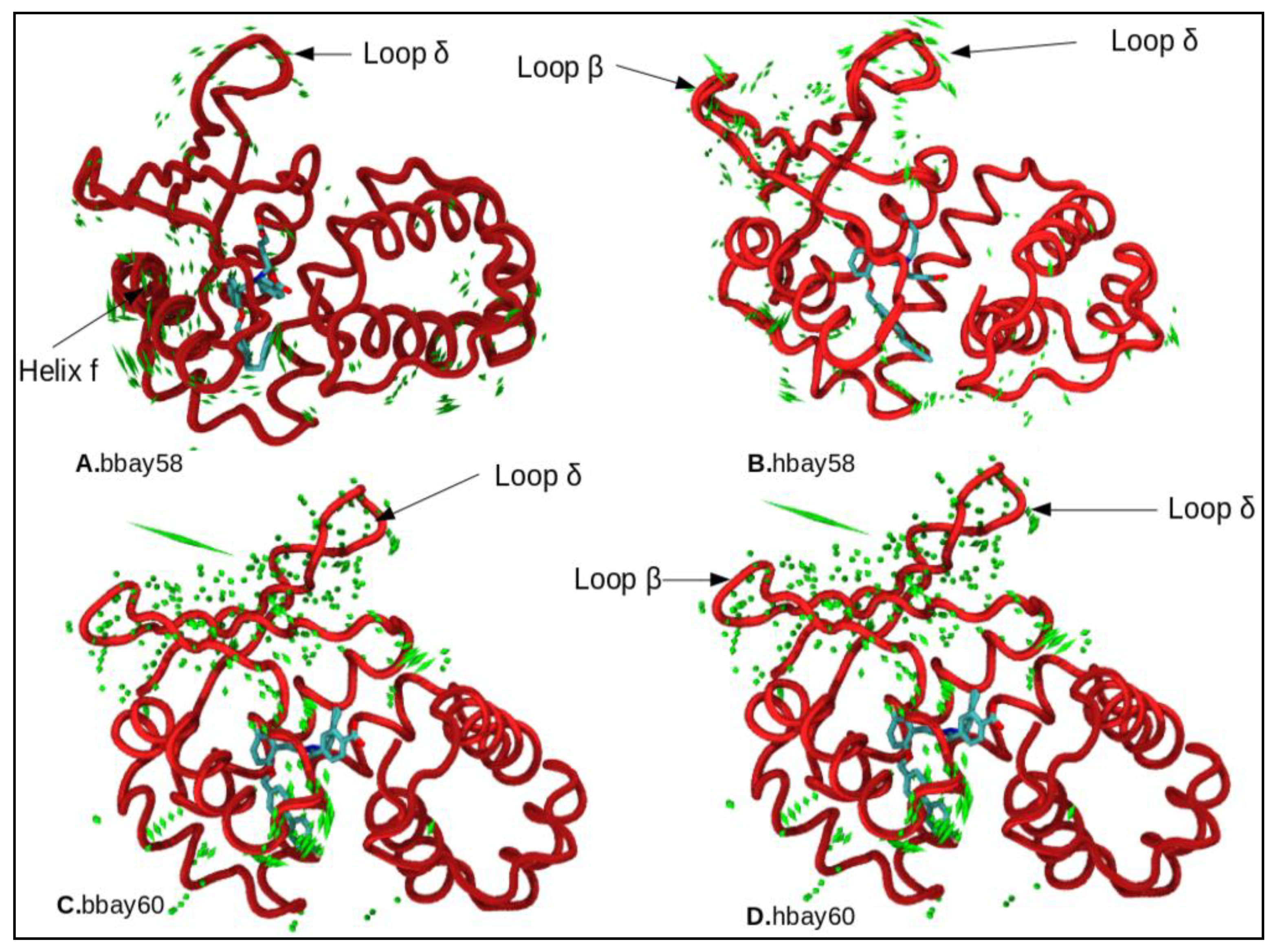
2.4. Principle Component Analysis
3. Materials and Methods
3.1. Comparative Modeling
3.2. sGC Activators Charge Derivation
3.3. Molecular Dynamics Simulation
3.4. Hydrogen Bond Occupancy
3.5. Principal Component Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Campbell, M.G.; Underbakke, E.S.; Potter, C.S.; Carragher, B.; Marletta, M.A. Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase. Proc. Natl. Acad. Sci. USA 2014, 111, 2960–2965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evgenov, O.V.; Pacher, P.; Schmidt, P.M.; Haskó, G.; Schmidt, H.H.; Stasch, J.P. NO-independent stimulators and activators of soluble guanylate cyclase: Discovery and therapeutic potential. Nat. Rev. Drug Discov. 2006, 5, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Underbakke, E.S.; Iavarone, A.T.; Chalmers, M.J.; Pascal, B.D.; Novick, S.; Griffin, P.R.; Marletta, M.A. Nitric Oxide-Induced Conformational Changes in Soluble Guanylate Cyclase. Struct. Des. 2014, 22, 602–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, B.; Koesling, D. cGMP signalling beyond nitric oxide. Trends Pharmacol. Sci. 2001, 22, 546–548. [Google Scholar] [CrossRef]
- Martin, E.; Berka, V.; Bogatenkova, E.; Murad, F.; Tsai, A.L. Ligand selectivity of soluble guanylyl cyclase: Effect of the hydrogen-bonding tyrosine in the distal heme pocket on binding of oxygen, nitric oxide, and carbon monoxide. J. Biol. Chem. 2006, 281, 27836–27845. [Google Scholar] [CrossRef] [PubMed]
- Montfort, W.R.; Wales, J.A.; Weichsel, A. Structure and Activation of Soluble Guanylyl Cyclase, the Nitric Oxide Sensor. Antioxid. Redox Signal. 2017, 26, 107–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nioche, P.; Berka, V.; Vipond, J.; Minton, N.; Tsai, A.L.; Raman, C.S. Femtomolar sensitivity of a NO sensor from Clostridium botulinum. Science 2004, 306, 1550. [Google Scholar] [CrossRef] [PubMed]
- Feelisch, M. The use of nitric oxide donors in pharmacological studies. Naunyn. Schmiedebergs Arch. Pharmacol. 1998, 358, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ignarro, L.J.; Harbison, R.G.; Wood, K.S.; Kadowitz, P.J. Dissimilarities between methylene blue and cyanide on relaxation and cyclic GMP formation in endothelium-intact intrapulmonary artery caused by nitrogen oxide-containing vasodilators and acetylcholine. J. Pharmacol. Exp. Ther. 1986, 236, 30–36. [Google Scholar] [PubMed]
- Stone, J.R.; Marletta, M.A. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry 1996, 35, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Martin, F.; Hahn, M.G.; Schaefer, M.; Stamler, J.S.; Stasch, J.P.; Van Den Akker, F. Insights into BAY60-2770 activation and S-nitrosylation-dependent desensitization of soluble guanylyl cyclase via crystal structures of homologous Nostoc H-NOX domain complexes. Biochemistry 2013, 52, 3601–3608. [Google Scholar] [CrossRef] [PubMed]
- Fouad, Y.M.; Yehia, R. Hepato-cardiac disorders. World J. Hepatol. 2014, 6, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Katsuki, S.; Arnold, W.; Mittal, C.; Murad, F. Stimulation of guanylate cyclase by sodium nitroprusside, nitroglycerin and nitric oxide in various tissue preparations and comparison to the effects of sodium azide and hydroxylamine. J. Cycl. Nucleotide Res. 1977, 3, 23–35. [Google Scholar]
- Stasch, J.P.; Becker, E.M.; Alonso-Alija, C.; Apeler, H.; Dembowsky, K.; Feurer, A.; Gerzer, R.; Minuth, T.; Perzborn, E.; Pleiß, U.; et al. NO-independent regulatory site on soluble guanylate cyclase. Nature 2001, 410. [Google Scholar] [CrossRef] [PubMed]
- Stasch, J.P.; Schmidt, P.; Alonso-Alija, C.; Apeler, H.; Dembowsky, K.; Haerter, M.; Heil, M.; Minuth, T.; Perzborn, E.; Pleiss, U.; et al. NO- and haem-independent activation of soluble guanylyl cyclase: Molecular basis and cardiovascular implications of a new pharmacological principle. Br. J. Pharmacol. 2002, 136, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Belik, J. Riociguat, an oral soluble guanylate cyclase stimulator for the treatment of pulmonary hypertension. Curr. Opin. Investig. Drugs 2009, 10, 971–979. [Google Scholar] [PubMed]
- Food and Drug Administration FDA Approves Adempas to Treat Pulmonary Hypertension. 2013. Available online: www.fda.gov/newsevents/newsroom/pressannouncements/ucm370866.htm (accessed on 29 April 2014).
- Khaybullina, D.; Patel, A.; Zerilli, T. Riociguat (adempas): A novel agent for the treatment of pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension. Pharm. Ther. 2014, 39, 749–758. [Google Scholar]
- Schindler, U.; Strobel, H.; Sch, K.; Linz, W.; Matthias, L.; Martorana, P.A.; Hartmut, R.; Schindler, P.W.; Busch, A.E.; Sohn, M.; et al. Biochemistry and Pharmacology of Novel Anthranilic Acid Derivatives Activating Heme-Oxidized Soluble Guanylyl Cyclase. Mol. Pharmacol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Nossaman, B.; Pankey, E.; Kadowitz, P. Stimulators and activators of soluble guanylate cyclase: Review and potential therapeutic indications. Crit. Care Res. Pract. 2012, 2012, 290805. [Google Scholar] [CrossRef] [PubMed]
- Schwemmer, M.; Bassenge, E. New approaches to overcome tolerance to nitrates. Cardiovasc. Drugs Ther. 2003, 17, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Baskaran, P.; Ma, X.; Dunten, P.W.; Schaefer, M.; Stasch, J.P.; Beuve, A.; Van Den Akker, F. Structure of cinaciguat (BAY58-2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J. Biol. Chem. 2010, 285, 22651–22657. [Google Scholar] [CrossRef] [PubMed]
- Lapp, H.; Mitrovic, V.; Franz, N.; Heuer, H.; Buerke, M.; Wolfertz, J.; Mueck, W.; Unger, S.; Wensing, G.; Frey, R. Cinaciguat (BAY 58-2667) improves cardiopulmonary hemodynamics in patients with acute decompensated heart failure. Circulation 2009, 119, 2781–2788. [Google Scholar] [CrossRef] [PubMed]
- Boerrigter, G.; Costello-Boerrigter, L.C.; Cataliotti, A.; Lapp, H.; Stasch, J.P.; Burnett, J.C., Jr. Targeting heme-oxidized soluble guanylate cyclase in experimental heart failure. Hypertension 2007, 49, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, E.; Semigran, M.J.; Nieminen, M.S.; Gheorghiade, M.; Agrawal, R.; Mitrovic, V.; Mebazaa, A. Cinaciguat, a soluble guanylate cyclase activator, unloads the heart but also causes hypotension in acute decompensated heart failure. Eur. Heart J. 2013, 34, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Tamargo, J.; Duarte, J.; Caballero, R.; Delpon, E. Cinaciguat, a soluble guanylate cyclase activator for the potential treatment of acute heart failure. Curr. Opin. Investig. Drugs 2010, 11, 1039–1047. [Google Scholar] [PubMed]
- Salloum, F.N.; Das, A.; Samidurai, A.; Hoke, N.N.; Chau, V.Q.; Ockaili, R.A.; Stasch, J.P.; Kukreja, R.C. Cinaciguat, a novel activator of soluble guanylate cyclase, protects against ischemia/reperfusion injury: Role of hydrogen sulfide. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1347–H1354. [Google Scholar] [CrossRef] [PubMed]
- Hausding, M.; Kopp, M.; Steven, S.; Schulz, E.; Stasch, J.P.; Munzel, T.; Daiber, A. Effect of soluble guanylyl cyclase activator and stimulator therapy on nitroglycerin-induced nitrate tolerance in rats. Vasc. Pharmacol. 2015, 71, 181–191. [Google Scholar] [Green Version]
- Tawa, M.; Shimosato, T.; Iwasaki, H.; Imamura, T.; Okamura, T. Different influences of extracellular and intracellular superoxide on relaxation through the NO/sGC/cGMP pathway in isolated rat iliac arteries. J. Cardiovasc. Pharmacol. 2015, 65, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, E.C.; Leiria, L.O.; Silva, F.H.; Mendes-Silverio, C.B.; Calmasini, F.B.; Davel, A.P.; Monica, F.Z.; De Nucci, G.; Antunes, E. Soluble guanylyl cyclase (sGC) degradation and impairment of nitric oxide-mediated responses in urethra from obese mice: Reversal by the sGC activator BAY60-2770. J. Pharmacol. Exp. Ther. 2014, 349, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Leiria, L.O.; Silva, F.H.; Davel, A.P.; Alexandre, E.C.; Calixto, M.C.; De Nucci, G.; Monica, F.Z.; Antunes, E. The soluble guanylyl cyclase activator BAY60-2770 ameliorates overactive bladder in obese mice. J. Urol. 2014, 191, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Silverio, C.B.; Leiria, L.O.; Morganti, R.P.; Anhe, G.F.; Marcondes, S.; Monica, F.Z.; De Nucci, G.; Antunes, E. Activation of haem-oxidized soluble guanylyl cyclase with BAY 60-2770 in human platelets lead to overstimulation of the cyclic GMP signaling pathway. PLoS ONE 2012, 7, e47223. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Koziol-White, C.J.; Asosingh, K.; Cheng, G.; Ruple, L.; Groneberg, D.; Friebe, A.; Comhair, S.A.; Stasch, J.P.; Panettieri, R.A., Jr.; et al. Soluble guanylate cyclase as an alternative target for bronchodilator therapy in asthma. Proc. Natl. Acad. Sci. USA 2016, 113, E2355–E2362. [Google Scholar] [CrossRef] [PubMed]
- Jabs, A.; Oelze, M.; Mikhed, Y.; Stamm, P.; Kroller-Schon, S.; Welschof, P.; Jansen, T.; Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [Green Version]
- Melo, F.; Sanchez, R.; Sali, A. Statistical potentials for fold assessment. Protein Sci. 2002, 11, 430–448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. ActaCrystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins Struct. Funct. Genet. 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Capece, L.; Estrin, D.A.; Marti, M.A. Dynamical characterization of the heme NO oxygen binding (HNOX) domain. Insight into soluble guanylate cyclase allosteric transition. Biochemistry 2008, 47, 9416–9427. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Kelley, C. Gnuplot4.4: An Interactive Plotting Program. 2010. 1986–1993. Available online: http//Sourceforge.Net/Projects/Gnuplot (accessed on 11 August 2018).
- Knorr, A.; Hirth-Dietrich, C.; Alonso-Alija, C.; Härter, M.; Hahn, M.; Keim, Y.; Wunder, F.; Stasch, J.-P. Nitric oxide-independent activation of soluble guanylate cyclase by BAY60-2770 in experimental liver fibrosis. Arzneimittelforschung 2008, 58, 71–80. [Google Scholar] [PubMed]
- Hoffmann, L.S.; Schmidt, P.M.; Keim, Y.; Hoffmann, C.; Schmidt, H.H.H.W.; Stasch, J.P. Fluorescence dequenching makes haem-free soluble guanylate cyclase detectable in living cells. PLoS ONE 2011, 6, e23596. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.-Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2006, 15, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System, Schrödinger LLC Wwwpymolorg. Version 1. 2002. Available online: http://www.pymol.org. doi:citeulike-article-id:240061 (accessed on 23 August 2006).
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Harvey, M.J.; De Fabritiis, G. An implementation of the smooth particle mesh Ewald method on GPU hardware. J. Chem. Theory Comput. 2009, 5, 2371–2377. [Google Scholar] [CrossRef] [PubMed]
- Gonnet, P. P-SHAKE: A quadratically convergent SHAKE in O (n2). J. Comput. Phys. 2007, 220, 740–750. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Turner, P.J. XMGRACE, Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| Complexes | Acceptor | Donor H | Donor | Percent of Frames of 50 ns Simulation Showing the Hydrogen Bond | Average Distance Å |
|---|---|---|---|---|---|
| bbay60 | V108@O | F112@H | F112@N | 24.58 | 2.88 |
| F112@O | L115@H | L115@N | 18.52 | 2.91 | |
| G109@O | F112@H | F112@N | 2.24 | 2.91 | |
| S111@O | F112@H | F112@N | 0.44 | 2.83 | |
| hbay60 | L108@O | Y112@H | Y112@N | 69.93 | 2.85 |
| Y112@O | M115@H | M115@N | 15.27 | 2.91 | |
| Y83@OH | Y112@HH | Y112@OH | 8.02 | 2.82 | |
| Y112@O | Y2@HH | Y2@OH | 0.20 | 2.77 | |
| bbay58 | V108@O | F112@H | F112@N | 37.43 | 2.87 |
| F112@O | L115@H | L115@N | 12.50 | 2.92 | |
| G109@O | F112@H | F112@N | 2.96 | 2.91 | |
| F112@O | Q114@H | Q114@N | 0.10 | 2.83 | |
| F112@O | Y2@HH | Y2@OH | 0.10 | 2.81 | |
| hbay58 | L108@O | Y112@H | Y112@N | 58.64 | 2.87 |
| Y112@OH | Y83@HH | Y83@OH | 8.59 | 2.83 | |
| Y112@O | M115@H | M115@N | 6.80 | 2.92 | |
| Y83@OH | Y112@H | Y112@OH | 1.63 | 2.81 | |
| V39@O | Y112@HH | Y112@OH | 1.56 | 2.79 | |
| Y112@OH | R40@HE | R40@NE | 0.21 | 2.90 | |
| Y112@OH | R40@HH12 | R40@NH1 | 0.10 | 2.89 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehan Khalid, R.; Tahir ul Qamar, M.; Maryam, A.; Ashique, A.; Anwar, F.; H. Geesi, M.; Siddiqi, A.R. Comparative Studies of the Dynamics Effects of BAY60-2770 and BAY58-2667 Binding with Human and Bacterial H-NOX Domains. Molecules 2018, 23, 2141. https://doi.org/10.3390/molecules23092141
Rehan Khalid R, Tahir ul Qamar M, Maryam A, Ashique A, Anwar F, H. Geesi M, Siddiqi AR. Comparative Studies of the Dynamics Effects of BAY60-2770 and BAY58-2667 Binding with Human and Bacterial H-NOX Domains. Molecules. 2018; 23(9):2141. https://doi.org/10.3390/molecules23092141
Chicago/Turabian StyleRehan Khalid, Rana, Muhammad Tahir ul Qamar, Arooma Maryam, Ayesha Ashique, Farooq Anwar, Mohammed H. Geesi, and Abdul Rauf Siddiqi. 2018. "Comparative Studies of the Dynamics Effects of BAY60-2770 and BAY58-2667 Binding with Human and Bacterial H-NOX Domains" Molecules 23, no. 9: 2141. https://doi.org/10.3390/molecules23092141






