Role of Resultant Dipole Moment in Mechanical Dissociation of Biological Complexes
Abstract
:1. Introduction
2. Results and Discussion
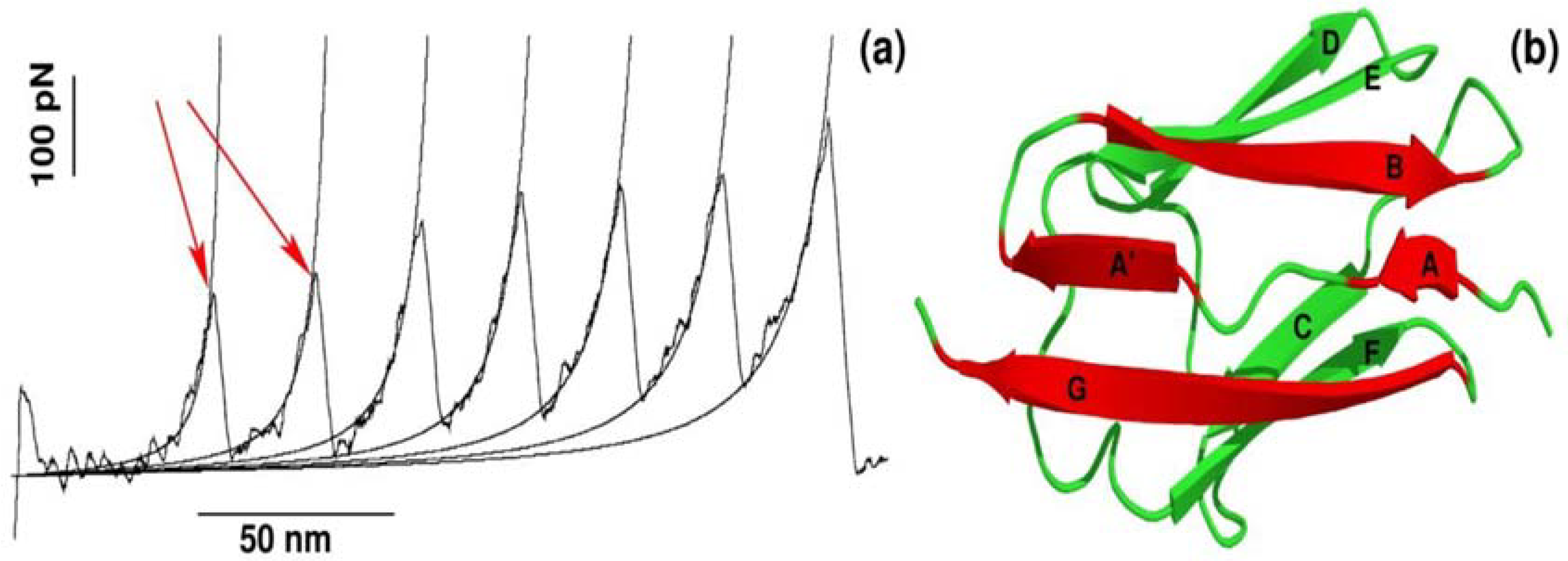
2.1. Assessing the Mechanical Stability of 2LLO Peptide-Protein Complex Using Steered Molecular Dynamics
2.2. Mechanical Stability Depends on the Pulling Direction of 2LLO Ligand-Receptor Complex
2.3. Robustness of Results Against Different Protein-Peptide Complexes
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Trott, O.; Olson, A.J. Software news and update autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- Wang, L.; Wu, Y.J.; Deng, Y.Q.; Kim, B.; Pierce, L.; Krilov, G.; Lupyan, D.; Robinson, S.; Dahlgren, M.K.; Greenwood, J.; et al. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J. Am. Chem. Soc. 2015, 137, 2695–2703. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.H.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, M.; Kurcinski, M.; Kouza, M.; Wieteska, L.; Debinski, A.; Kolinski, A.; Kmiecik, S. Modeling of protein-peptide interactions using the cabs-dock web server for binding site search and flexible docking. Methods 2016, 93, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The cluspro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. Cabs-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta flexpepdock web server-high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef] [PubMed]
- Bruce, N.J.; Ganotra, G.K.; Kokh, D.B.; Sadiq, S.K.; Wade, R.C. New approaches for computing ligand-receptor binding kinetics. Curr. Opin. Struct. Biol. 2018, 49, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, M.S.; Mai, B.K. Steered molecular dynamics-a promising tool for drug design. Curr. Bioinform. 2012, 7, 342–351. [Google Scholar]
- Rydzewski, J.; Nowak, W. Ligand diffusion in proteins via enhanced sampling in molecular dynamics. Phys. Life Rev. 2017, 22–23, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Van Vuong, Q.; Nguyen, T.T.; Li, M.S. A new method for navigating optimal direction for pulling ligand from binding pocket: Application to ranking binding affinity by steered molecular dynamics. J. Chem. Inf. Model. 2015, 55, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Grubmuller, H.; Heymann, B.; Tavan, P. Ligand binding: Molecular mechanics calculation of the streptavidin biotin rupture force. Science 1996, 271, 997–999. [Google Scholar] [CrossRef] [PubMed]
- Bernetti, M.; Cavalli, A.; Mollica, L. Protein-ligand (un)binding kinetics as a new paradigm for drug discovery at the crossroad between experiments and modelling. MedChemComm 2017, 8, 534–550. [Google Scholar] [CrossRef]
- Colizzi, F.; Perozzo, R.; Scapozza, L.; Recanatini, M.; Cavalli, A. Single-molecule pulling simulations can discern active from inactive enzyme inhibitors. J. Am. Chem. Soc. 2010, 132, 7361–7371. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.F.; Li, H.X.; Wang, X.C. A self-adaptive steered molecular dynamics method based on minimization of stretching force reveals the binding affinity of protein-ligand complexes. Molecules 2015, 20, 19236–19251. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Hsin, J.; Sotomayor, M.; Comellas, G.; Schulten, K. Discovery through the computational microscope. Structure 2009, 17, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Hu, C.K.; Li, M.S.; Kolinski, A. A structure-based model fails to probe the mechanical unfolding pathways of the titin i27 domain. J. Chem. Phys. 2013, 139, 065103. [Google Scholar] [CrossRef] [PubMed]
- Lichter, S.; Rafferty, B.; Flohr, Z.; Martini, A. Protein high-force pulling simulations yield low-force results. PLoS ONE 2012, 7, e34781. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Isralewitz, B.; Krammer, A.; Vogel, V.; Schulten, K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 1998, 75, 662–671. [Google Scholar] [CrossRef]
- Lemkul, J.A.; Bevan, D.R. Assessing the stability of alzheimer’s amyloid protofibrils using molecular dynamics. J. Phys. Chem. B 2010, 114, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Co, N.T.; Li, M.S.; Kmiecik, S.; Kolinski, A.; Kloczkowski, A.; Buhimschi, I.A. Kinetics and mechanical stability of the fibril state control fibril formation time of polypeptide chains: A computational study. J. Chem. Phys. 2018, 148, 215106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.Y. An inhomogeneous model of protein dielectric properties: Intrinsic polarizabilities of amino acids. J. Chem. Phys. 2002, 116, 9359–9363. [Google Scholar] [CrossRef]
- Brockwell, D.J.; Paci, E.; Zinober, R.C.; Beddard, G.S.; Olmsted, P.D.; Smith, D.A.; Perham, R.N.; Radford, S.E. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat. Struct. Biol. 2003, 10, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Paci, E.; Hummer, G.; Dudko, O.K. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J. Phys. Chem. B 2008, 112, 5968–5976. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by afm. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Marszalek, P.E.; Lu, H.; Li, H.B.; Carrion-Vazquez, M.; Oberhauser, A.F.; Schulten, K.; Fernandez, J.M. Mechanical unfolding intermediates in titin modules. Nature 1999, 402, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Kotamarthi, H.C.; Sharma, R.; Ainavarapu, R.K. Single-molecule studies on polysumo proteins reveal their mechanical flexibility. Biophys. J. 2013, 104, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Li, M.S. Biomolecules under mechanical force. Phys. Rep. 2010, 486, 1–74. [Google Scholar] [CrossRef]
- Kouza, M.; Lan, P.D.; Gabovich, A.M.; Kolinski, A.; Li, M.S. Switch from thermal to force-driven pathways of protein refolding. J. Chem. Phys. 2017, 146, 135101. [Google Scholar] [CrossRef] [PubMed]
- Glyakina, A.V.; Likhachev, I.V.; Balabaev, N.K.; Galzitskaya, O.V. Right- and left-handed three-helix proteins. Ii. Similarity and differences in mechanical unfolding of proteins. Proteins 2014, 82, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Hu, C.K.; Zung, H.; Li, M.S. Protein mechanical unfolding: Importance of non-native interactions. J. Chem. Phys. 2009, 131, 215103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.S. Ligand migration and steered molecular dynamics in drug discovery: Comment on “ligand diffusion in proteins via enhanced sampling in molecular dynamic” by jakub rydzewski and wieslaw nowak. Phys. Life Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Schulten, K. Steered molecular dynamics simulation of conformational changes of immunoglobulin domain i27 interpret atomic force microscopy observations. Chem. Phys. 1999, 247, 141–153. [Google Scholar] [CrossRef]
- Fowler, S.B.; Best, R.B.; Herrera, J.L.T.; Rutherford, T.J.; Steward, A.; Paci, E.; Karplus, M.; Clarke, J. Mechanical unfolding of a titin Ig domain: Structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J. Mol. Biol. 2002, 322, 841–849. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.G.; Sacks, D.B.; Ames, J.B. Structural basis for Ca2+-induced activation and dimerization of estrogen receptor alpha by calmodulin. J. Biol. Chem. 2012, 287, 9336–9344. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Arad-Haase, G.; Chuartzman, S.G.; Dagan, S.; Nevo, R.; Kouza, M.; Binh, K.M.; Hung, T.N.; Li, M.S.; Reich, Z. Mechanical unfolding of acylphosphatase studied by single-molecule force spectroscopy and md simulations. Biophys. J. 2010, 99, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Klimov, D.K.; Thirumalai, D. Stretching single-domain proteins: Phase diagram and kinetics of force-induced unfolding. Proc. Natl. Acad. Sci. USA 1999, 96, 6166–6170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irie, K.; Nakatsu, T.; Mitsuoka, K.; Miyazawa, A.; Sobue, K.; Hiroaki, Y.; Doi, T.; Fujiyoshi, Y.; Kato, H. Crystal structure of the homer 1 family conserved region reveals the interaction between the evh1 domain and own proline-rich motif. J. Mol. Biol. 2002, 318, 1117–1126. [Google Scholar] [CrossRef]
- Wu, J.W.; Cocina, A.E.; Chai, J.J.; Hay, B.A.; Shi, Y.G. Structural analysis of a functional diap1 fragment bound to grim and hid peptides. Mol. Cell. 2001, 8, 95–104. [Google Scholar] [CrossRef]
- Scott, W.R.P.; Hunenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Kruger, P.; van Gunsteren, W.F. The gromos biomolecular simulation program package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules (vol 117, pg 5179, 1995). J. Am. Chem. Soc. 1996, 118, 2309. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction models for water in relation to protein hydration. Intermol. Forces 1981, 14, 331–442. [Google Scholar]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Wabik, J.; Kmiecik, S.; Gront, D.; Kouza, M.; Kolinski, A. Combining coarse-grained protein models with replica-exchange all-atom molecular dynamics. Int. J. Mol. Sci. 2013, 14, 9893–9905. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Banerji, A.; Kolinski, A.; Buhimschi, I.A.; Kloczkowski, A. Oligomerization of fvflm peptides and their ability to inhibit beta amyloid peptides aggregation: Consideration as a possible model. Phys. Chem. Chem. Phys. 2017, 19, 2990–2999. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Hansmann, U.H.E. Velocity scaling for optimizing replica exchange molecular dynamics. J. Chem. Phys. 2011, 134, 044124. [Google Scholar] [CrossRef] [PubMed]
- Kouza, M.; Co, N.T.; Nguyen, P.H.; Kolinski, A.; Li, M.S. Preformed template fluctuations promote fibril formation: Insights from lattice and all-atom models. J. Chem. Phys. 2015, 142, 145104. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh ewald—An n.Log(n) method for ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. Lincs: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Peplowski, L.; Sikora, M.; Nowak, W.; Cieplak, M. Molecular jamming-the cystine slipknot mechanical clamp in all-atom simulations. J. Chem. Phys. 2011, 134. [Google Scholar] [CrossRef] [PubMed]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein binding pocket dynamics. Accounts Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef] [PubMed]
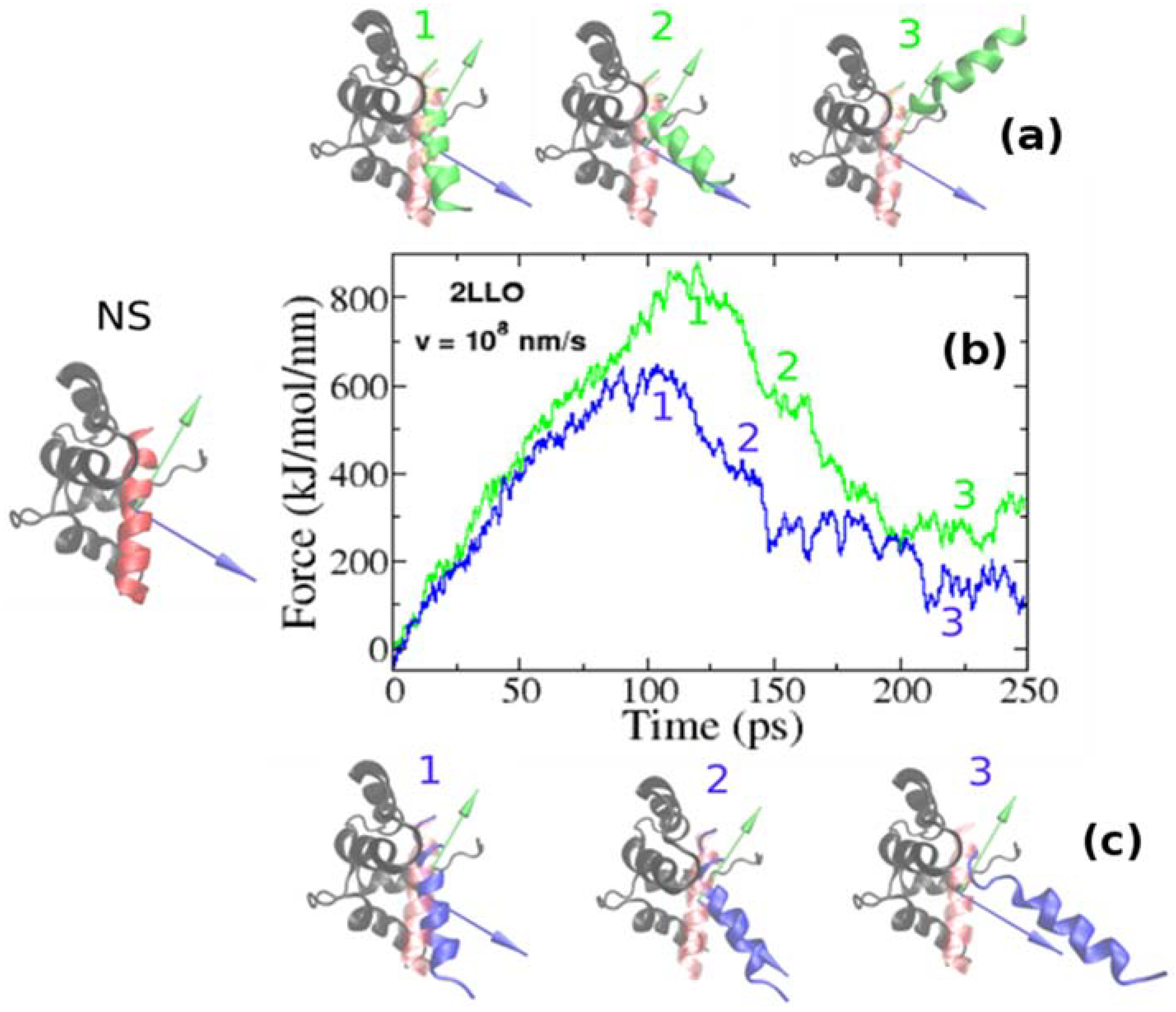
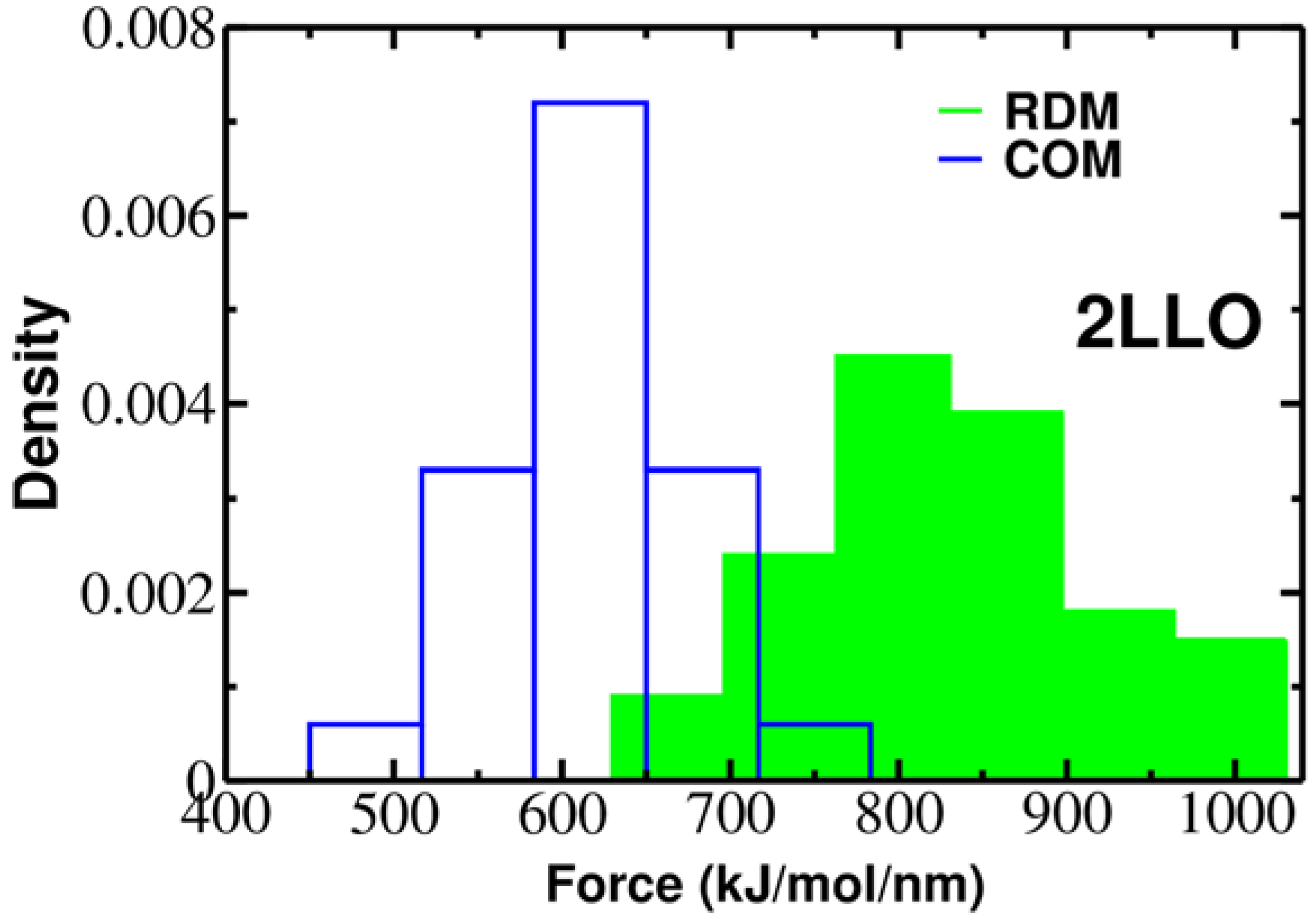
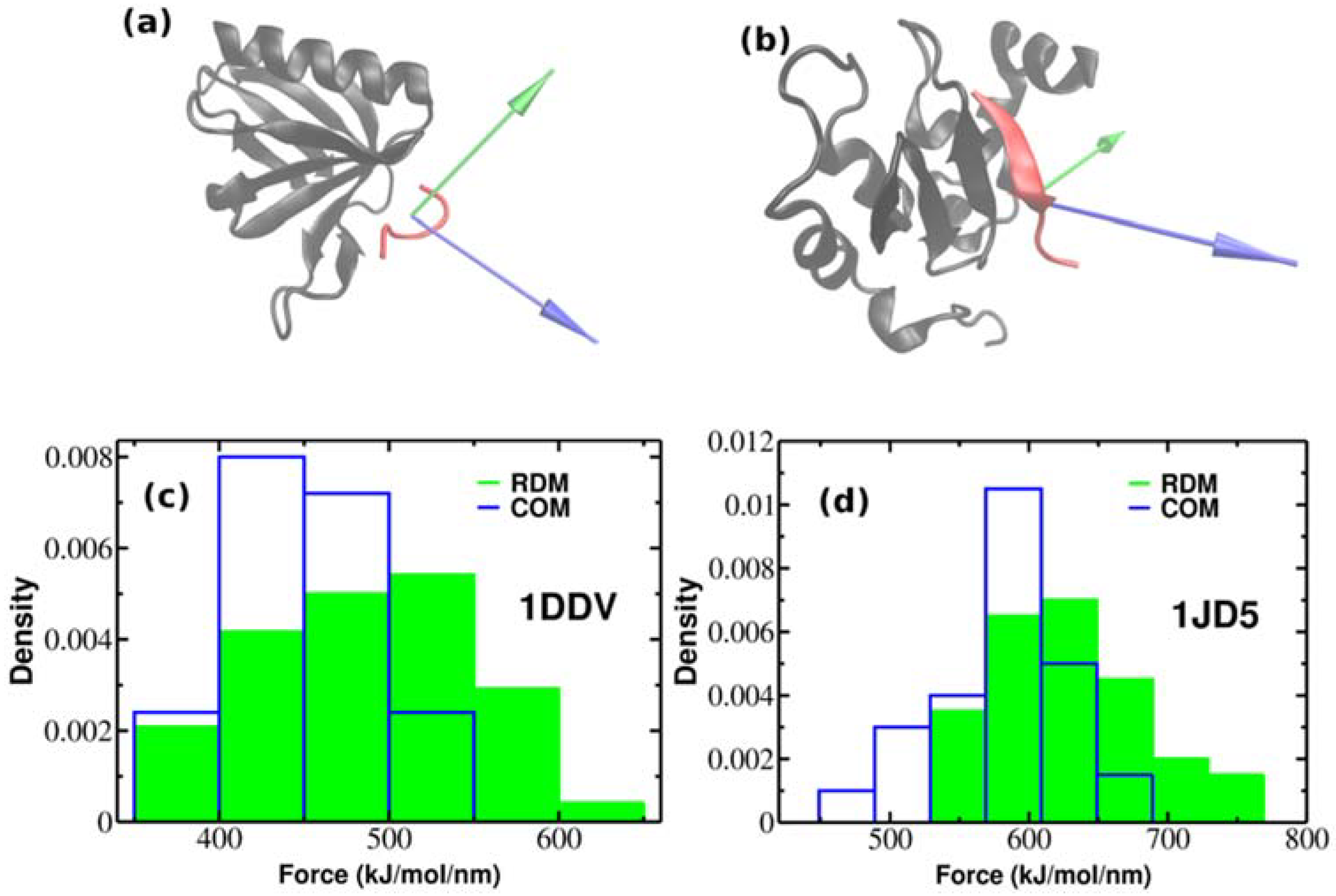
| PDB Code of the Protein-Peptide Complex | Identified Protein Residues Involved in Protein-Peptide Interactions | RDM Vector | COM Vector | Force (kJ/mol/nm) RDM COM | |
|---|---|---|---|---|---|
| 2LLO | 7–21, 25–29, 31–40, 43, 47–58, 61–65, 67–80 | 0.144i + 0.983j − 0.11k | 0.193i + 0.068j + 0.979k | 828.5 ± 118.7 | 613.5 ± 56.4 |
| 1DDV | 10–16, 22–26, 30–31, 69–76,87–92, 96, 109 | −0.636i + 0.111j + 0.764k | −0.799i + 0.454j − 0.395k | 486.1 ± 53.6 | 432.4 ± 42.1 |
| 1JD5 | 219–220, 242, 252–257, 259–263, 265–279, 282–283, 285–290, 311, 314–315, 317–318 | 0.226i + 0.182j − 0.957k | −0.77i − 0.416j − 0.484k | 773.9 ± 149.4 | 595.9 ± 55.1 |
| PDB ID of The Protein-Peptide Complex | Protein | Peptide | ||
|---|---|---|---|---|
| Length | Class | Length | Class | |
| 2LLO | 80 | α/β (34/2%) | 19 | α (84%) |
| 1DDV | 104 | α/β (13/45%) | 6 | unstructured |
| 1JD5 | 105 | α/β (41/7%) | 8 | β (40%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouza, M.; Banerji, A.; Kolinski, A.; Buhimschi, I.; Kloczkowski, A. Role of Resultant Dipole Moment in Mechanical Dissociation of Biological Complexes. Molecules 2018, 23, 1995. https://doi.org/10.3390/molecules23081995
Kouza M, Banerji A, Kolinski A, Buhimschi I, Kloczkowski A. Role of Resultant Dipole Moment in Mechanical Dissociation of Biological Complexes. Molecules. 2018; 23(8):1995. https://doi.org/10.3390/molecules23081995
Chicago/Turabian StyleKouza, Maksim, Anirban Banerji, Andrzej Kolinski, Irina Buhimschi, and Andrzej Kloczkowski. 2018. "Role of Resultant Dipole Moment in Mechanical Dissociation of Biological Complexes" Molecules 23, no. 8: 1995. https://doi.org/10.3390/molecules23081995





