Environmentally Friendly Approach to Knoevenagel Condensation of Rhodanine in Choline Chloride: Urea Deep Eutectic Solvent and QSAR Studies on Their Antioxidant Activity
Abstract
:1. Introduction
2. Results and Discussion
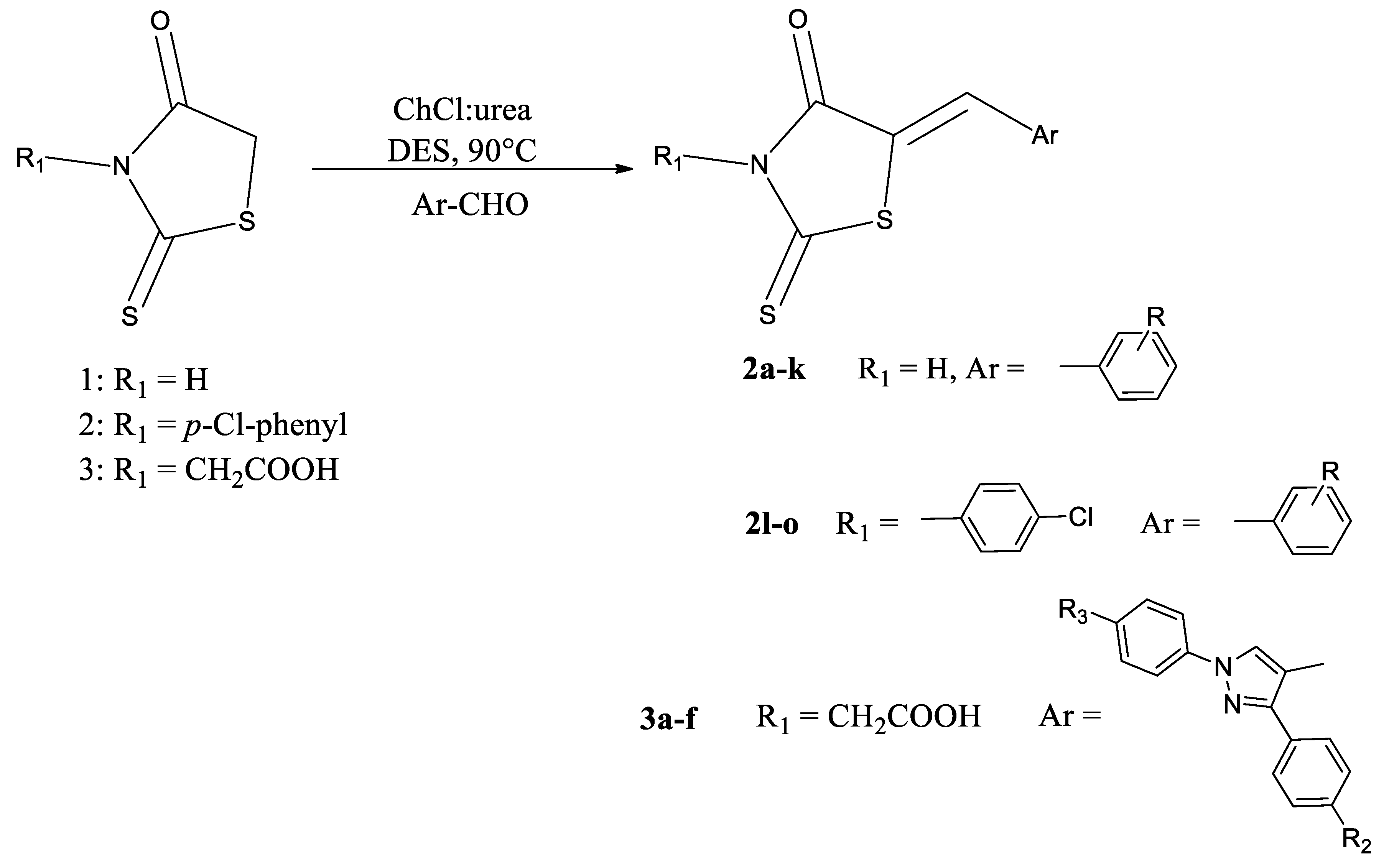
2.1. Synthesis
2.2. Antioxidant Activity
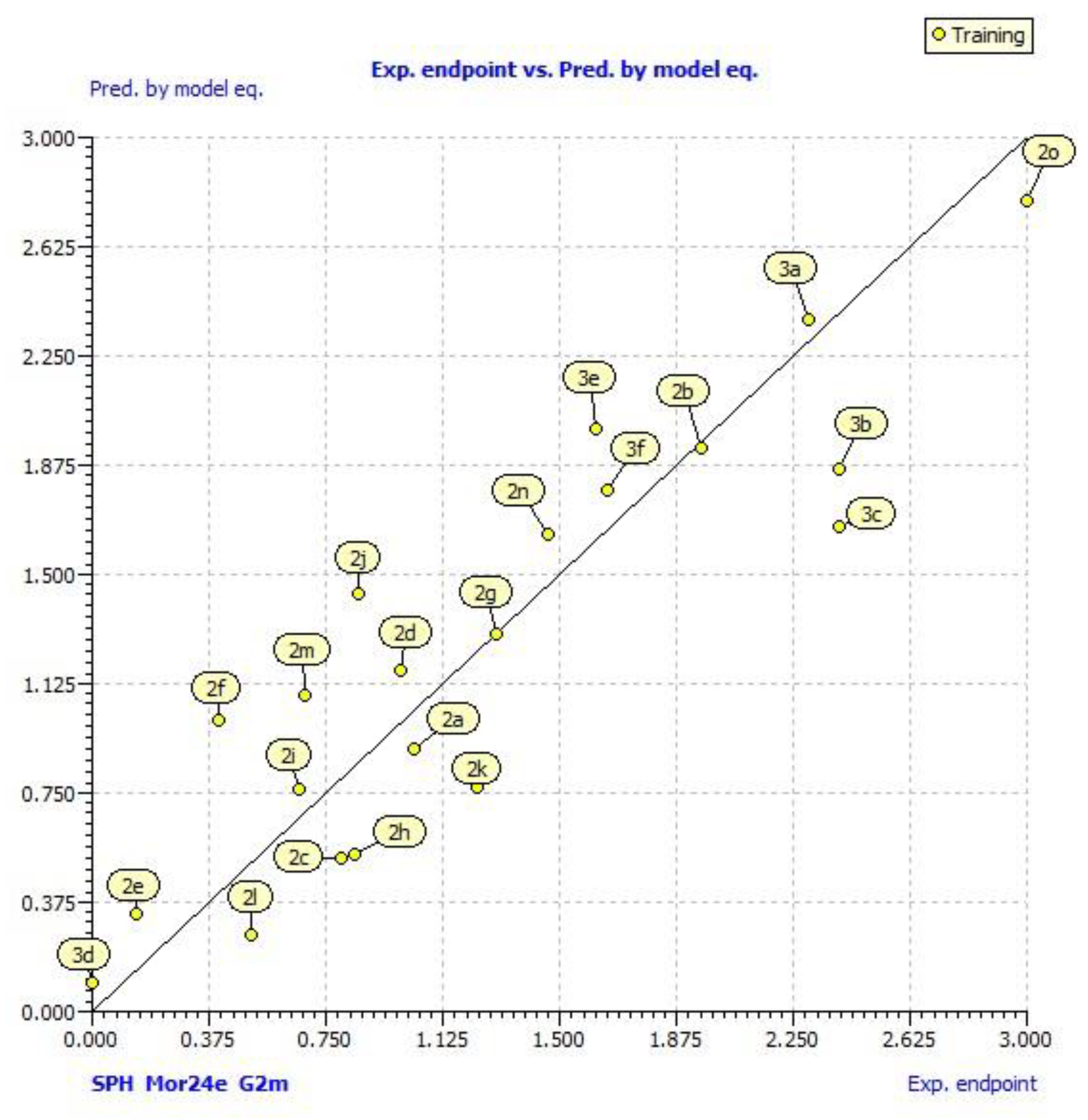
2.3. QSAR Models
2.4. Molecular Docking Study
3. Materials and Methods
3.1. General
3.2. DES Preparation
3.3. Knoevenagel Condensation in Choline Chloride:Urea (1:2) DES
3.4. Antioxidant Activity
3.5. QSAR Method
3.5.1. Data Set
3.5.2. Descriptor Calculation
3.5.3. Regression Analysis and Validation of Models
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Azizi, N.; Dezfooli, S.; Khajeh, M.; Hashemi, M.M. Efficient deep eutectic solvents catalyzed synthesis of pyran and benzopyran derivatives. J. Mol. Liq. 2013, 186, 76–80. [Google Scholar] [CrossRef]
- Dai, Y.; van Spronsen, J.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta 2013, 766, 61–68. [Google Scholar] [CrossRef] [PubMed]
- García, A.; Rodríguez-Juan, E.; Rodríguez-Gutiérrez, G.; Rios, J.J.; Fernández-Bolaños, J. Extraction of phenolic compounds from virgin olive oil by deep eutectic solvents (DESs). Food Chem. 2016, 197, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Harishkumar, H.N.; Mahadevan, K.M.; Kumar, C.K.; Satyanarayan, N.D. A facile, choline chloride/urea catalyzed solid phase synthesis of coumarins via Knoevenagel condensation. Org. Commun. 2011, 4, 26. [Google Scholar]
- Khandelwal, S.; Tailor, Y.K.; Kumar, M. Deep eutectic solvents (DESs) as eco-friendly and sustainable solvent/catalyst systems in organic transformations. J. Mol. Liq. 2016, 215, 345–386. [Google Scholar] [CrossRef]
- Lobo, H.R.; Singh, B.S.; Shankarling, G.S. Bio-compatible eutectic mixture for multi-component synthesis: A valuable acidic catalyst for synthesis of novel 2,3-dihydroquinazolin-4(1H)-one derivatives. Catal. Commun. 2012, 27, 179–183. [Google Scholar] [CrossRef]
- More, P.A.; Gadilohar, B.L.; Shankarling, G.S. In Situ Generated Cetyltrimethylammonium Bisulphate in Choline Chloride—Urea Deep Eutectic Solvent: A Novel Catalytic System for One Pot Synthesis of 1,3,4-Oxadiazole. Catal. Lett. 2014, 8, 1393–1398. [Google Scholar] [CrossRef]
- Pednekar, S.; Bhalerao, R.; Ghadge, N. One-pot multi-component synthesis of 1,4-dihydropyridine derivatives in biocompatible deep eutectic solvents. J. Chem. Sci. 2013, 125, 615–621. [Google Scholar] [CrossRef]
- Rajawat, A.; Khandelwal, S.; Kumar, M. Deep eutectic solvent promoted efficient and environmentally benign four-component domino protocol for synthesis of spirooxindoles. RSC Adv. 2014, 4, 5105–5112. [Google Scholar] [CrossRef]
- Zhu, A.; Jiang, T.; Han, B.; Zhang, J.; Xie, Y.; Ma, X. Supported choline chloride/urea as a heterogeneous catalyst for chemical fixation of carbon dioxide to cyclic carbonates. Green Chem. 2007, 9, 169–172. [Google Scholar] [CrossRef]
- De Santi, V.; Cardellini, F.; Brinchi, L.; Germani, R. Novel Brønsted acidic deep eutectic solvent as reaction media for esterification of carboxylic acid with alcohols. Tetrahedron Lett. 2012, 53, 5151–5155. [Google Scholar] [CrossRef]
- Martínez, R.; Berbegal, L.; Guillena, G.; Ramón, D.J. Bio-renewable enantioselective aldol reaction in natural deep eutectic solvents. Green Chem. 2016, 18, 1724–1730. [Google Scholar] [CrossRef] [Green Version]
- Phadtare, S.B.; Shankarling, G.S. Halogenation reactions in biodegradable solvent: Efficient bromination of substituted 1-aminoanthra-9,10-quinone in deep eutectic solvent (choline chloride: Urea). Green Chem. 2010, 12, 458–462. [Google Scholar] [CrossRef]
- Amić, A.; Molnar, M. An Improved and Efficient N-acetylation of Amines Using Choline Chloride Based Deep Eutectic Solvents. Org. Prep. Proced. Int. 2017, 49, 249–257. [Google Scholar] [CrossRef]
- Molnar, M.; Klenkar, J.; Tarnai, T. Eco-friendly rapid synthesis of 3-substituted-2-thioxo-2,3-dihydroquinazolin-4(1H)-ones in choline chloride based deep eutectic solvent. Synth. Commun. 2017, 47, 1040–1045. [Google Scholar] [CrossRef]
- Molnar, M.; Komar, M.; Brahmbhatt, H.; Babić, J.; Jokić, S.; Rastija, V. Deep Eutectic Solvents as Convenient Media for Synthesis of Novel Coumarinyl Schiff Bases and Their QSAR Studies. Molecules 2017, 22, 1482. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzipour, F.; Tavakol, H. The synthesis of coumarin derivatives using choline chloride/zinc chloride as a deep eutectic solvent. J. Iran. Chem. Soc. 2016, 13, 149–153. [Google Scholar] [CrossRef]
- Yarovenko, V.N.; Nikitina, A.S.; Zavarzin, I.V.; Krayushkin, M.M.; Kovalenko, L.V. Synthesis of 2-thioxo-1,3-thiazolidin-4-one derivatives. Russ. Chem. Bull. 2007, 56, 1624–1630. [Google Scholar] [CrossRef]
- Radi, M.; Botta, L.; Casaluce, G.; Bernardini, M.; Botta, M. Practical one-pot two-step protocol for the microwave-assisted synthesis of highly functionalized rhodanine derivatives. J. Comb. Chem. 2010, 12, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Klein, C.D. Aqueous microwave-assisted one-pot synthesis of N-substituted rhodanines. Tetrahedron Lett. 2012, 53, 5197–5201. [Google Scholar] [CrossRef]
- Azizi, N.; Hasani, M.; Khajeh, M.; Edrisi, M. A straightforward and sustainable one-pot, four-component synthesis of rhodanine derivatives. Tetrahedron Lett. 2015, 56, 1189–1192. [Google Scholar] [CrossRef]
- Sandhu, J.S. Ultrasound-assisted synthesis of 2,4-thiazolidinedione and rhodanine derivatives catalyzed by task-specific ionic liquid: [TMG][Lac]. Org. Med. Chem. Lett. 2013, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Zervosen, A.; Lu, W.-P.; Chen, Z.; White, R.E.; Demuth, T.P.; Frère, J.-M. Interactions between penicillin-binding proteins (PBPs) and two novel classes of PBP inhibitors, arylalkylidene rhodanines and arylalkylidene iminothiazolidin-4-ones. Antimicrob. Agents Chemother. 2004, 48, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Dayam, R.; Sanchez, T.; Clement, O.; Shoemaker, R.; Sei, S.; Neamati, N. β-Diketo Acid Pharmacophore Hypothesis. 1. Discovery of a Novel Class of HIV-1 Integrase Inhibitors. J. Med. Chem. 2005, 48, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Takasu, K.; Pudhom, K.; Kaiser, M.; Brun, R.; Ihara, M. Synthesis and Antimalarial Efficacy of Aza-Fused Rhodacyanines in vitro and in the P. berghei Mouse Model. J. Med. Chem. 2006, 49, 4795–4798. [Google Scholar] [CrossRef] [PubMed]
- Sortino, M.; Delgado, P.; Juárez, S.; Quiroga, J.; Abonía, R.; Insuasty, B.; Nogueras, M.; Rodero, L.; Garibotto, F.M.; Enriz, R.D.; et al. Synthesis and antifungal activity of (Z)-5-arylidenerhodanines. Bioorg. Med. Chem. 2007, 15, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fan, Y.-H.; Natarajan, A.; Guo, Y.; Iyasere, J.; Harbinski, F.; Luus, L.; Christ, W.; Aktas, H.; Halperin, J.A. Synthesis and biological evaluation of thiazolidine-2,4-dione and 2,4-thione derivatives as inhibitors of translation initiation. Bioorg. Med. Chem. Lett. 2004, 14, 5401–5405. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pang, Q. Oxidative stress-associated protein tyrosine kinases and phosphatases in Fanconi anemia. Antioxid. Redox Signal. 2014, 20, 2290–2301. [Google Scholar] [CrossRef] [PubMed]
- Masand, V.H.; Mahajan, D.T.; Nazeruddin, G.M.; Hadda, T.B.; Rastija, V.; Alfeefy, A.M. Effect of information leakage and method of splitting (rational and random) on external predictive ability and behavior of different statistical parameters of QSAR model. Med. Chem. Res. 2015, 24, 1241–1264. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; John Wiley & Sons: Hoboken, NJ, USA, 2008; ISBN 978-3-527-61311-3. [Google Scholar]
- Devinyak, O.; Havrylyuk, D.; Lesyk, R. 3D-MoRSE descriptors explained. J. Mol. Graph. Model. 2014, 54, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, R.; Gramatica, P. 3D-modelling and Prediction by WHIM Descriptors. Part 6. Application of WHIM Descriptors in QSAR Studies. Quant. Struct.-Act. Relatsh. 1997, 16, 120–125. [Google Scholar] [CrossRef]
- Sicheri, F.; Moarefi, I.; Kuriyan, J. Crystal structure of the Src family tyrosine kinase Hck. Nature 1997, 385, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; O’Donoghue, R.J.J.; Ernst, M. Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget 2015, 6, 15752–15771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subhedar, D.D.; Shaikh, M.H.; Nawale, L.; Yeware, A.; Sarkar, D.; Shingate, B.B. [Et3NH][HSO4] catalyzed efficient synthesis of 5-arylidene-rhodanine conjugates and their antitubercular activity. Res. Chem. Intermed. 2016, 42, 6607–6626. [Google Scholar] [CrossRef]
- Swathi, T.; Srinivas, M. Green condensation reaction of aromatic aldehydes with rhodanine catalyzed by alum under microwave irradiation. Der Pharma Chem. 2015, 5, 100–104. [Google Scholar]
- Bhatti, R.S.; Singh, M.; Sandhu, J.S. Microwave Enhanced Synthesis OF 5-Arylidene Rhodanine under solvent free conditions using bismuth trichloride as a promoter. Rasayan J. Chem. 2008, 5, 738–742. [Google Scholar]
- Kumar, D.; Narwal, S.; Sandhu, J.S. Catalyst-free synthesis of highly biologically active 5-arylidene rhodanine and 2,4-thiazolidinedione derivatives using aldonitrones in polyethylene glycol. Int. J. Med. Chem. 2013, 2013, 273534. [Google Scholar] [CrossRef] [PubMed]
- Šarkanj, B.; Molnar, M.; Čačić, M.; Gille, L. 4-Methyl-7-hydroxycoumarin antifungal and antioxidant activity enhancement by substitution with thiosemicarbazide and thiazolidinone moieties. Food Chem. 2013, 139, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Hocquet, A.; Langgård, M. An Evaluation of the MM+ Force Field. J. Mol. Med. 1998, 4, 94–112. [Google Scholar] [CrossRef]
- Tetko, I.V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V.A.; Radchenko, E.V.; Zefirov, N.S.; Makarenko, A.S.; et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aided Mol. Des. 2005, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- ADMEWORKS: Fujitsu Kyushu Systems. Available online: http://www.fujitsu.com/jp/group/kyushu/en/solutions/industry/lifescience/admeworks/ (accessed on 15 May 2018).
- Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. QSARINS: A new software for the development, analysis, and validation of QSAR MLR models. J. Comput. Chem. 2013, 34, 2121–2132. [Google Scholar] [CrossRef]
- Wiley Online Library. Principles of QSAR models validation: Internal and external-Gramatica. QSAR & Combinatorial Science, 2007. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/qsar.200610151 (accessed on 15 May 2018).
- Hsu, K.-C.; Chen, Y.-F.; Lin, S.-R.; Yang, J.-M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011, 12 (Suppl. S1), S33. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of all compounds are available from the authors. |



| Solvent | Yield (%) |
|---|---|
| choline chloride:urea | 78 |
| 1st recycle | 78 |
| 2nd recycle | 74 |
| 3rd recycle | 76 |
 | ||||||||||||||
| Cmp. | R1 | %DPPH | pDPPH | |||||||||||
| 1 | H | 3.6 | 1.444 | |||||||||||
| 2 | p-Cl-phenyl | 94.4 | 0.025 | |||||||||||
| 3 | CH2COOH | 23.9 | 0.622 | |||||||||||
 | ||||||||||||||
| Cmp. | Ar | R1 | % DPPH | pDPPH | Cmp. | Ar | R1 | % DPPH | pDPPH | |||||
| 2a | 2-OCH3-phenyl | H | 9.2 | 1.036 | 2i | 4-pyridyl | H | 21.5 | 0.668 | |||||
| 2b | 4-N(CH3)2-phenyl | H | 1.1 | 1.959 | 2j | 2,5-(OCH3)2-phenyl | H | 13.9 | 0.857 | |||||
| 2c | 3-OCH3-phenyl | H | 15.7 | 0.804 | 2k | 4-OH-phenyl | H | 5.8 | 1.237 | |||||
| 2d | 3,4,5-(OCH3)3-phenyl | H | 10.1 | 0.996 | 2l | 3-OH-4-OCH3-phenyl | 4-Cl-phenyl | 30.8 | 0.511 | |||||
| 2e | 3,4-(OH)2-phenyl | H | 71.2 | 0.148 | 2m | 4-N(CH3)2-phenyl | 4-Cl-phenyl | 20.6 | 0.686 | |||||
| 2f | 3-OCH3-4-OH-phenyl | H | 39.2 | 0.407 | 2n | 4-OH-phenyl | 4-Cl-phenyl | 3.4 | 1.469 | |||||
| 2g | 2-OH-phenyl | H | 5.0 | 1.301 | 2o | 2,5-(OCH3)2-phenyl | 4-Cl-phenyl | 0.1 | 3 | |||||
| 2h | phenyl | H | 14.2 | 0.848 | ||||||||||
 | ||||||||||||||
| Cmp. | R1 | R2 | R3 | % DPPH | pDPPH | |||||||||
| 3a | CH2COOH | -CH3 | -CH3 | 0.5 | 2.301 | |||||||||
| 3b | CH2COOH | -F | -CH3 | 0.4 | 2.398 | |||||||||
| 3c | CH2COOH | -OCH3 | -CH3 | 0.4 | 2.398 | |||||||||
| 3d | CH2COOH | -Cl | -CH3 | 0 | 0 | |||||||||
| 3e | CH2COOH | -Br | -CH3 | 2.4 | 1.62 | |||||||||
| 3f | CH2COOH | -I | -CH3 | 2.2 | 1.658 | |||||||||
| Complex | Total Energy/kcal·mol−1 | van der Waals Interaction | H Bond | Elec |
|---|---|---|---|---|
| 2HCK + 2e | −77.1535 | −53.5621 | −23.5914 | 0 |
| H Bond | Energy | van der Waals interaction | Energy | |
| H-M-MET-341 | −8.5 | V-S-LEU-273 | −8.07 | |
| H-M-SER-345 | −3.5 | V-S-VAL-281 | −4.13 | |
| H-S-SER-345 | −3.46 | V-S-PHE-340 | −4.79 | |
| H-S-ASP-348 | −6.60 | V-M-GLY-344 | −5.96 | |
| V-M-SER-345 | −5.28 | |||
| V-S-LEU-393 | −4.10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molnar, M.; Brahmbhatt, H.; Rastija, V.; Pavić, V.; Komar, M.; Karnaš, M.; Babić, J. Environmentally Friendly Approach to Knoevenagel Condensation of Rhodanine in Choline Chloride: Urea Deep Eutectic Solvent and QSAR Studies on Their Antioxidant Activity. Molecules 2018, 23, 1897. https://doi.org/10.3390/molecules23081897
Molnar M, Brahmbhatt H, Rastija V, Pavić V, Komar M, Karnaš M, Babić J. Environmentally Friendly Approach to Knoevenagel Condensation of Rhodanine in Choline Chloride: Urea Deep Eutectic Solvent and QSAR Studies on Their Antioxidant Activity. Molecules. 2018; 23(8):1897. https://doi.org/10.3390/molecules23081897
Chicago/Turabian StyleMolnar, Maja, Harshad Brahmbhatt, Vesna Rastija, Valentina Pavić, Mario Komar, Maja Karnaš, and Jurislav Babić. 2018. "Environmentally Friendly Approach to Knoevenagel Condensation of Rhodanine in Choline Chloride: Urea Deep Eutectic Solvent and QSAR Studies on Their Antioxidant Activity" Molecules 23, no. 8: 1897. https://doi.org/10.3390/molecules23081897








