Decomposing the Mechanism of Qishen Granules in the Treatment of Heart Failure by a Quantitative Pathway Analysis Method
Abstract
:1. Introduction
2. Results
2.1. Echocardiographic Assessment of Left Ventricular Function
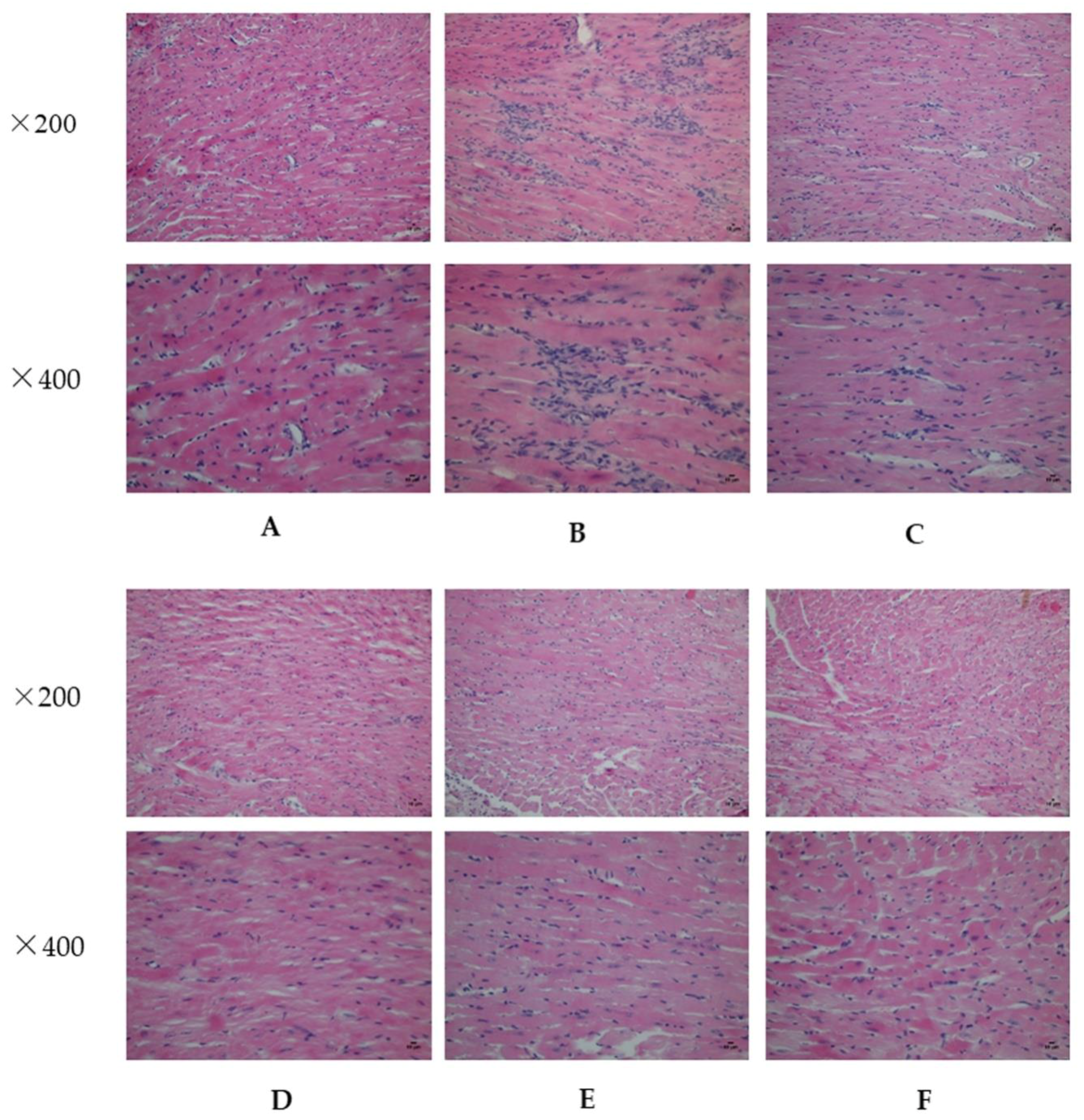
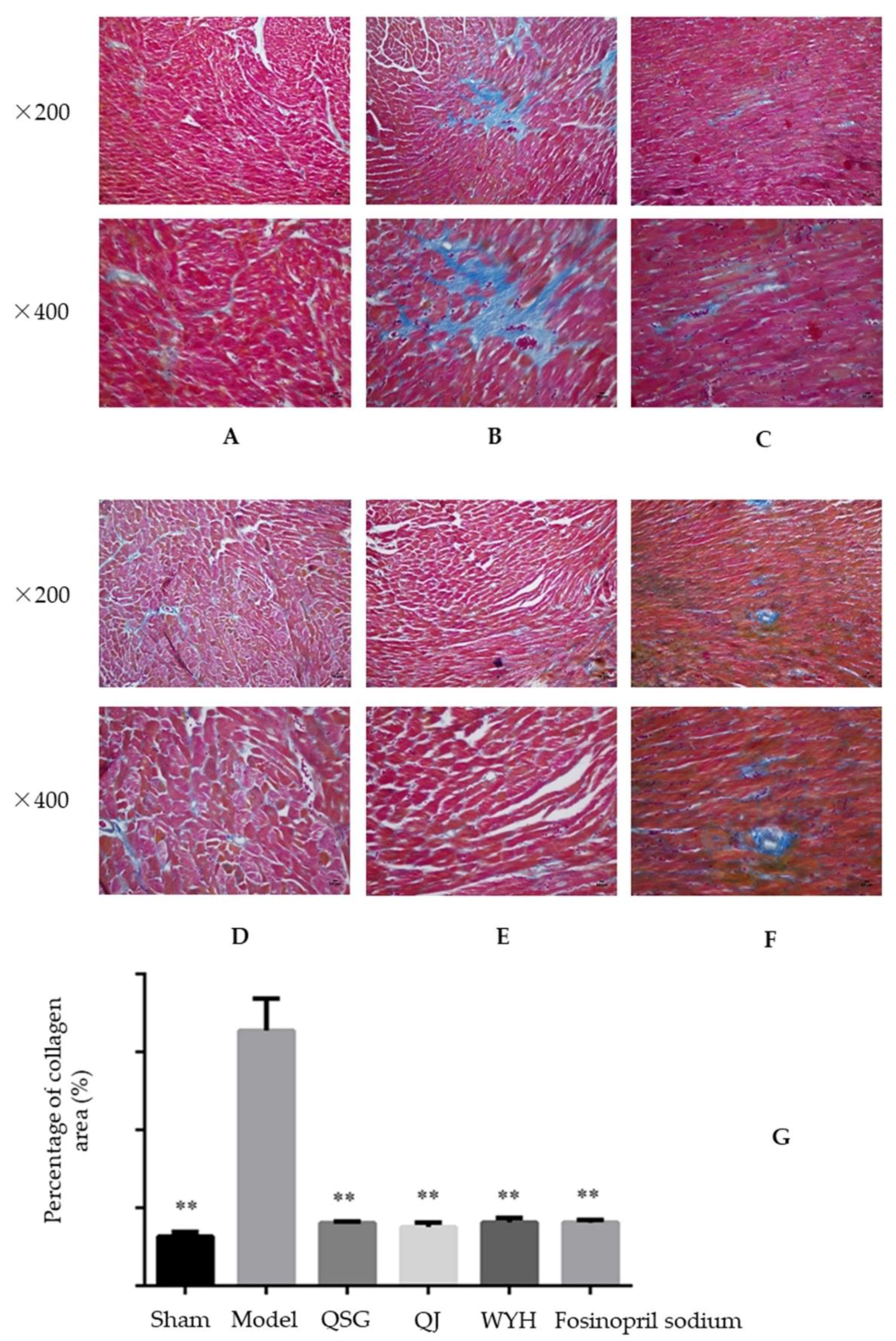
2.2. Analysis of Myocardial Hypertrophy
2.3. Transcriptome Results
2.3.1. Differential Expression Analysis
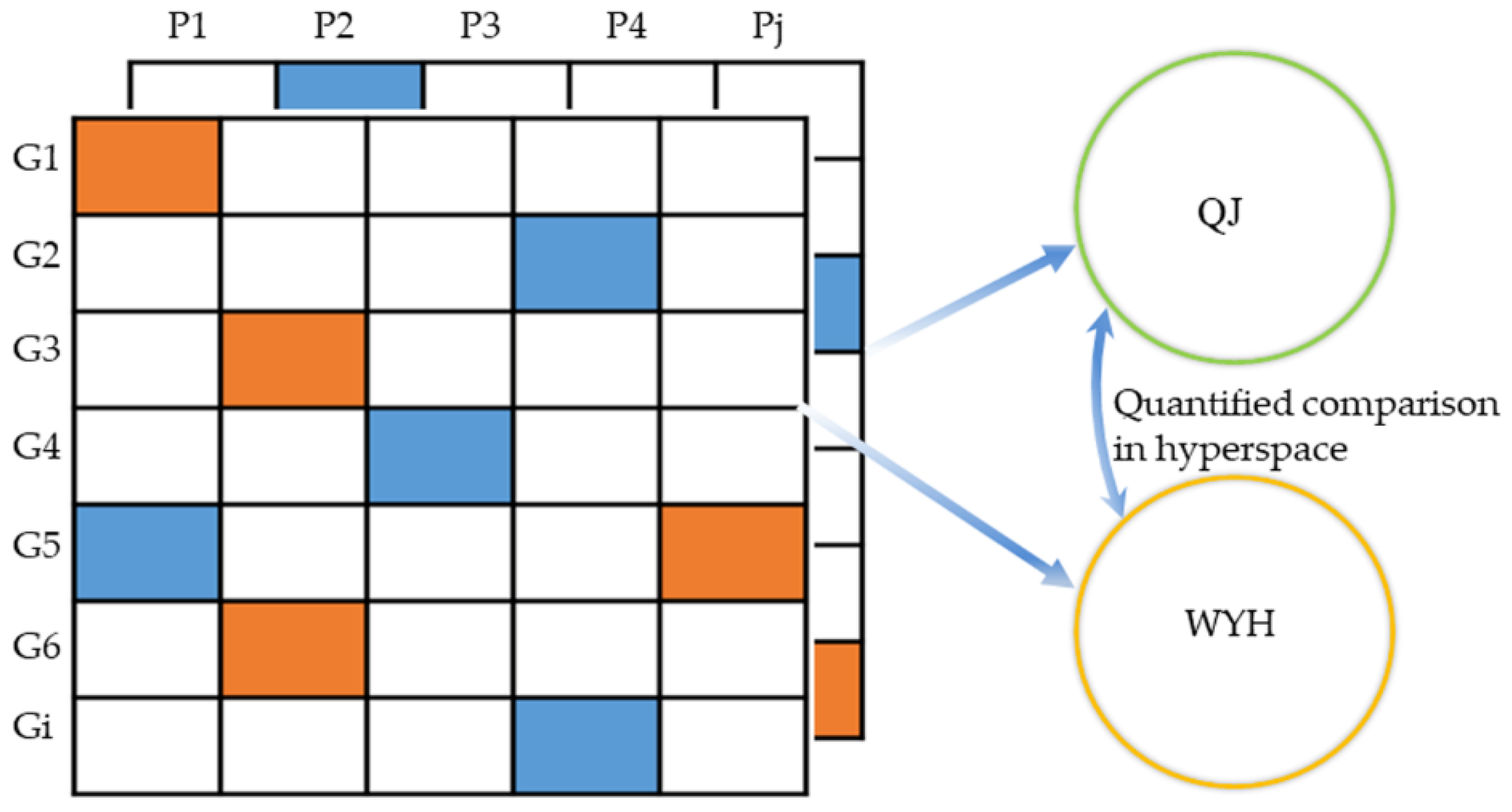
2.3.2. Quantitative Pathway Analysis
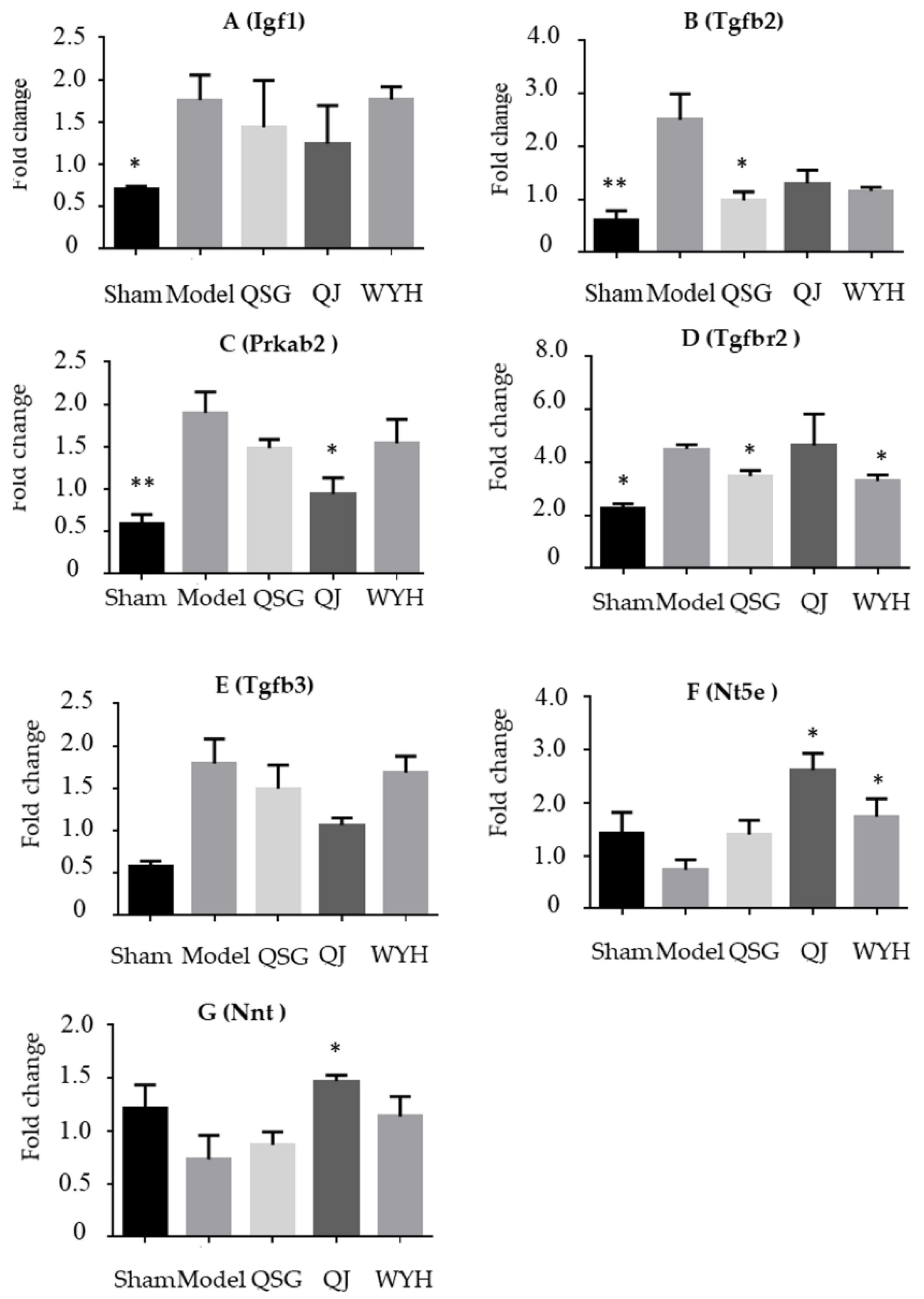
2.4. Validation of the Key Genes in HF-Related Pathways
2.5. Western Blot Detection of the Nnt and Nt5e Protein Levels
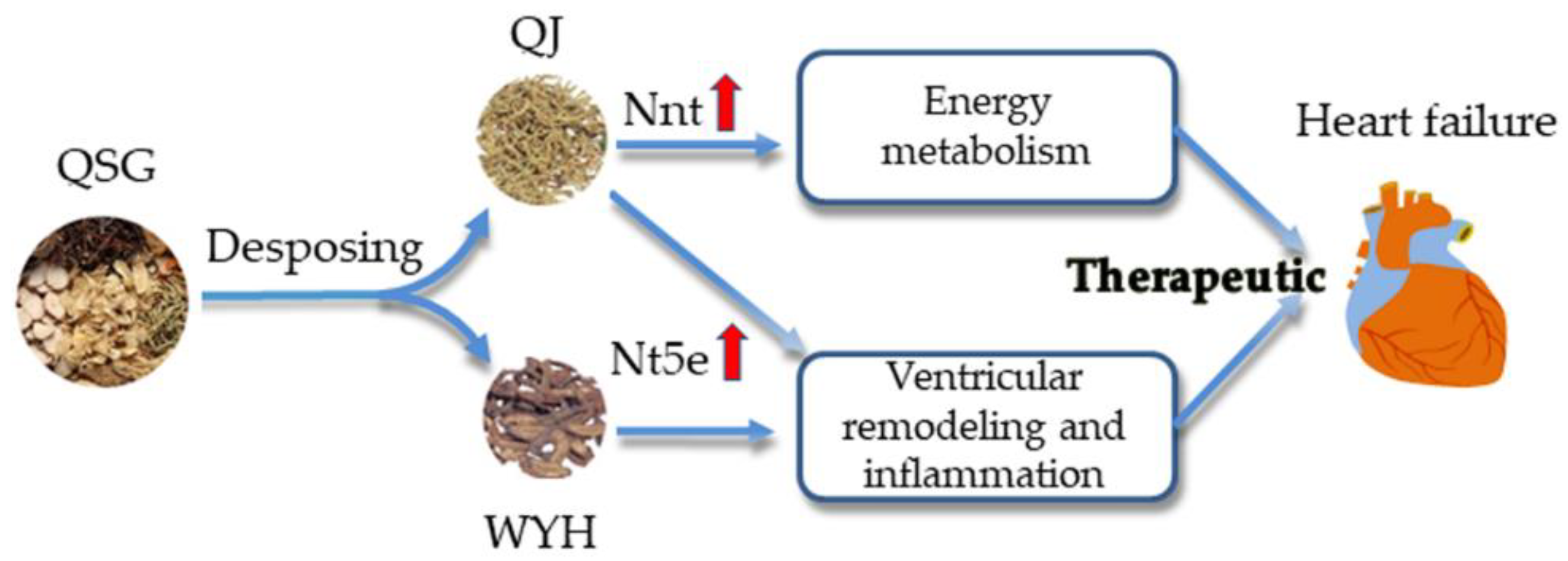
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. TAC Model Establishment and Grouping
4.3. Preparation and Determination of the Dose of QSG, QJ, WYH, and Fosinopril Sodium Tablets
4.4. Echocardiographic Assessment of Left Ventricular Function
4.5. Histopathological Examination
4.6. Microarray Analysis
4.6.1. RNA Extraction, Amplification, Labeling, and Hybridization
4.6.2. Differential Expression Analysis
4.6.3. Quantitative Pathway Analysis
4.7. Real-Time Fluorescence Quantitative PCR for Verifying the Results of the Trancriptome
4.8. Measurement of Indicators by Western Blot
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, L.; Yan, K.P.; Liu, X.C.; Wang, W.; Li, C.; Li, M.; Qiu, C.G. Valsartan regulates TGF-beta/Smads and TGF-beta/p38 pathways through LncRNA CHRF to improve doxorubicin-induced heart failure. Arch. Pharm. Res. 2017, 41, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Roger, V.L. Epidemiology of heart failure. Circ. Res. 2013, 113, 646–659. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, F. Epidemiology and prognosis of heart failure. Herz 2015, 40, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.; Coats, A.J.; Falk, V.; Gonzalez-Juanatey, J.R.; Harjola, V.P.; Jankowska, E.A.; et al. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: The task force for the diagnosis and treatment of acute and chronic heart failure of the European society of cardiology (ESC) Developed with the special contribution of the heart failure association (HFA) of the esc. Eur. Heart J. 2016, 18, 891–975. [Google Scholar]
- Shimizu, I.; Minamino, T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell Cardiol. 2016, 97, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Zagorianou, A.; Marougkas, M.; Drakos, S.G.; Diakos, N.; Konstantopoulos, P.; Perrea, D.N.; Anastasiou-Nana, M.; Malliaras, K. The effect of long-term amiodarone administration on myocardial fibrosis and evolution of left ventricular remodeling in a porcine model of ischemic cardiomyopathy. SpringerPlus 2016, 5, 1568. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Dorn, G.W., 2nd. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 2001, 63, 391–426. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Olson, E.N. Cardiac hypertrophy: The good, the bad, and the ugly. Annu. Rev. Physiol. 2003, 65, 45–79. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.P.; Davidson, S.M.; Townsend, P.A. Molecular regulation of cardiac hypertrophy. Int. J. Biochem. Cell Biol. 2008, 40, 2023–2039. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, Y.; Lin, W.K.; Zhang, Y.; Liu, W.; Huang, K.; Terrar, D.A.; Solaro, R.J.; Wang, X.; Ke, Y.; et al. Inhibition of angiotensin ii-induced cardiac hypertrophy and associated ventricular arrhythmias by a p21 activated kinase 1 bioactive peptide. PLoS ONE 2014, 9, e101974. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Li, C.; Lu, L.; Zhang, Q.; Zhu, R.; Wang, W. A review of Chinese herbal medicine for the treatment of chronic heart failure. Curr. Pharm. Des. 2017, 23, 5115–5124. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, Y.; Ren, W.; Guo, S.; Wei, W. Discussion on the significance of treating heart failure with clearing heat and removing toxicity from the inflammation. Chin. J. Tradit. Chin. Med. Pharm. 2017, 11, 4901–4905. [Google Scholar]
- Guo, S.; Li, P.; Fu, B.; Chuo, W.; Gao, K.; Zhang, W.; Wang, J.; Chen, J.; Wang, W. Systems-biology dissection of mechanisms and chemical basis of herbal formula in treating chronic myocardial ischemia. Pharmacol. Res. 2016, 114, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, Y.; Sun, J.; Li, C.; Song, Y.; Li, J.; Tu, P.; Zhao, Y. Simultaneous determination of twenty-five compounds in rat plasma using ultra-high performance liquid chromatography-polarity switching tandem mass spectrometry and its application to a pharmacokinetic study. Molecules 2017, 22, 1853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, C.; Ouyang, Y.; Yu, J.; Guo, S.; Liu, Z.; Li, D.; Han, J.; Wang, W. Cardioprotective effects of Qishenyiqi mediated by angiotensin ii type 1 receptor blockade and enhancing angiotensin-converting enzyme 2. Evid. Based Complement. Altern. Med. 2012, 2012, 978127. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Li, C.; Li, D.; Ouyang, Y.; Yu, J.; Guo, S.; He, F.; Wang, W. Drug target prediction based on the herbs components: The study on the multitargets pharmacological mechanism of Qishenkeli acting on the coronary heart disease. Evid. Based Complement. Altern. Med. 2012, 2012, 698531. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Yu, X.; Fan, X.; Wang, W.; Guo, S. Qishen Granule Regulated the Fas/Fasl Apoptotic Pathway in Myocardial Tissuess of Mice with Transversse Aortic Constriction. Chin. J. Integr. Med. Cardio 2017, 6, 667–670. [Google Scholar]
- Wang, Y.; Lin, W.; Li, C.; Singhal, S.; Jain, G.; Zhu, L.; Lu, L.; Zhu, R.; Wang, W. Multipronged therapeutic effects of Chinese herbal medicine Qishenyiqi in the treatment of acute myocardial infarction. Front. Pharmacol. 2017, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Li, C.; Wang, Q.; Lu, L.; Zhang, Q.; Zhang, Y.; Zhang, N.; Wang, Y.; Wang, W. QSKL protects against myocardial apoptosis on heart failure via PI3K/Akt-p53 signaling pathway. Sci. Rep. 2017, 7, 16986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, K.; Wang, Q.; Li, C.; Zeng, Z.; Wang, Y.; Wang, W. Effect of QSKL on MAPK and RHOA pathways in a rat model of heart failure. Evid. Based Complement. Altern. Med. 2017, 2017, 3903898. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, J.; Zhang, W.; Fu, B.; Wang, W. Transcriptome inference and systems approaches to polypharmacology and drug discovery in herbal medicine. J. Ethnopharmacol. 2017, 195, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, J.; Wang, Q.; Zhang, Y.; Zhang, N.; Lu, L.; Wu, Y.; Zhang, Q.; Wang, W.; Wang, Y.; et al. Qishen granules inhibit myocardial inflammation injury through regulating arachidonic acid metabolism. Sci. Rep. 2016, 6, 36949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.H.; Li, C.; Wang, Q.Y.; Zhang, Q.; Zhang, Y.; Meng, H.; Wang, Y.; Wang, W. Cardioprotective effects of Qishen Granule () on sarcoplasmic reticulum Ca(2+) handling in heart failure rats. Chin. J. Integr. Med. 2017, 23, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Zhu, Y.; Fan, X.H.; Zhang, B.L. Efficacy-oriented compatibility for component-based Chinese medicine. Acta Pharmacol. Sin. 2015, 36, 654–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, B.A.; Asif, A.R.; Schnelle, M.; Qasim, M.; Khadjeh, S.; Lbik, D.; Schott, P.; Hasenfuss, G.; Toischer, K. Proteomic analysis of short-term preload-induced eccentric cardiac hypertrophy. J. Transl. Med. 2016, 14, 149. [Google Scholar] [CrossRef] [PubMed]
- Albracht, S.P.; Meijer, A.J.; Rydstrom, J. Mammalian NADH:Ubiquinone oxidoreductase (Complex I) and nicotinamide nucleotide transhydrogenase (Nnt) together regulate the mitochondrial production of H(2)O(2)–implications for their role in disease, especially cancer. J. Bioenerg. Biomembr. 2011, 43, 541–564. [Google Scholar] [CrossRef] [PubMed]
- Fisher-Wellman, K.H.; Lin, C.T.; Ryan, T.E.; Reese, L.R.; Gilliam, L.A.; Cathey, B.L.; Lark, D.S.; Smith, C.D.; Muoio, D.M.; Neufer, P.D. Pyruvate dehydrogenase complex and nicotinamide nucleotide transhydrogenase constitute an energy-consuming redox circuit. Biochem. J. 2015, 467, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Energy metabolism in heart failure. J. Physiol. 2004, 555, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doenst, T.; Nguyen, T.D.; Abel, E.D. Cardiac metabolism in heart failure: Implications beyond ATP production. Circ. Res. 2013, 113, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Shirazi, L.F.; Bissett, J.; Romeo, F.; Mehta, J.L. Role of inflammation in heart failure. Curr. Atheroscler. Rep. 2017, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Carlson, D.; Sun, Y.; Ma, L.; Wolf, S.E.; Minei, J.P.; Zang, Q.S. Mitochondrial ROS induces cardiac inflammation via a pathway through mtDNA damage in a pneumonia-related sepsis model. PLoS ONE 2015, 10, e0139416. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.; Zazueta, C.; Aguilera-Aguirre, L. Oxidative stress and inflammation in cardiovascular disease. Oxid. Med. Cell Longev. 2017, 2017, 5853238. [Google Scholar] [CrossRef] [PubMed]
- Dick, S.A.; Epelman, S. Chronic heart failure and inflammation: What do we really know? Circ. Res. 2016, 119, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Bonner, F.; Borg, N.; Jacoby, C.; Temme, S.; Ding, Z.; Flogel, U.; Schrader, J. Ecto-5′-nucleotidase on immune cells protects from adverse cardiac remodeling. Circ. Res. 2013, 113, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Sassi, Y.; Ahles, A.; Truong, D.J.; Baqi, Y.; Lee, S.Y.; Husse, B.; Hulot, J.S.; Foinquinos, A.; Thum, T.; Muller, C.E.; et al. Cardiac myocyte-secreted camp exerts paracrine action via adenosine receptor activation. J. Clin. Investig. 2014, 124, 5385–5397. [Google Scholar] [CrossRef] [PubMed]
- Koon, C.M.; Zhang, X.; Chen, W.; Chu, E.S.; San Lau, C.B.; Wang, Y.X. Black blood t1rho MR imaging may diagnose early stage liver fibrosis: A proof-of-principle study with rat biliary duct ligation model. Quant. Imaging Med. Surg. 2016, 6, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Kuster, B.; Culhane, A.C.; Gholami, A.M. A multivariate approach to the integration of multi-omics datasets. BMC Bioinform. 2014, 15, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |

| Group | N | LVIDd (mm) | LVIDs (mm) | LVPWd (mm) | LVPWs (mm) | LVAWd (mm) | LVAWs (mm) | LVEDV (μL) | LVESV (μL) | LVM (mg) | EF (%) | FS (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sham | 7 | 4.28 ± 0.09 ** | 3.12 ± 0.08 ** | 0.54 ± 0.03 ** | 0.54 ± 0.02 ** | 0.51 ± 0.03 ** | 0.51 ± 0.02 ** | 82.59 ± 4.02 ** | 38.82 ± 2.43 ** | 77.23 ± 6.34 ** | 52.59 ± 3.02 ** | 26.98 ± 2.00 ** |
| Model | 10 | 4.85 ± 0.13 ▲▲ | 4.14 ± 0.18 ▲▲ | 0.76 ± 0.02 ▲▲ | 0.73 ± 0.02 ▲▲ | 0.67 ± 0.02 ▲▲ | 0.7 ± 0.02 ▲▲ | 111.41 ± 6.77 ▲▲ | 77.95 ± 8.03 ▲▲ | 142.83 ± 9.43 ▲▲ | 31.51 ± 3.29 ▲▲ | 15.05 ± 1.69 ▲▲ |
| QSG | 8 | 4.57 ± 0.10 | 3.63 ± 0.13 * | 0.52 ± 0.01 ** | 0.51 ± 0.02 ** | 0.48 ± 0.01 ** | 0.49 ± 0.02 ** | 96.11 ± 4.67 | 56.43 ± 4.69 * | 82.00 ± 4.04 ** | 41.73 ± 2.95 * | 20.51 ± 1.62 * |
| WYH | 8 | 4.53 ± 0.15 * | 3.47 ± 0.15 ** | 0.57 ± 0.15 ** | 0.63 ± 0.05 * | 0.57 ± 0.16 **,# | 0.56 ± 0.02 **,# | 94.96 ± 7.36 * | 51.01 ± 5.18 ** | 94.27 ± 6.35 ** | 46.67 ± 2.78 ** | 23.39 ± 1.68 ** |
| QJ | 8 | 4.39 ± 0.12 * | 3.42 ± 0.11 ** | 0.54 ± 0.02 ** | 0.57 ± 0.02 ** | 0.58 ± 0.02 *,## | 0.57 ± 0.03 **,# | 88.24 ± 5.65 * | 48.89 ± 3.92 ** | 88.62 ± 5.82 ** | 44.66 ± 2.52 ** | 22.12 ± 1.49 * |
| Fosinopril sodium | 8 | 4.36 ± 0.17 * | 3.32 ± 0.23 ** | 0.62 ± 0.02 ** | 0.61 ± 0.02 * | 0.57 ± 0.03 ** | 0.62 ± 0.02 * | 86.30 ± 2.80 * | 45.11 ± 2.62 ** | 93.77 ± 4.61 ** | 47.92 ± 1.85 ** | 22.99 ± 1.12 ** |
| Entrez ID | Gene Symbol | QJ Mean Expression | Fold Change | p-Value | WYH Mean Expression | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|
| 16000 | Igf1 | 64.23 | 0.490 | 0.013↓* | 77.62 | 0.595 | 0.057↓ |
| 108097 | Prkab2 | 467.22 | 0.457 | 0.040↓* | 515.14 | 0.459 | 0.045↓* |
| 21809 | Tgfb3 | 468.04 | 0.237 | 0.021↓* | 658.34 | 0.327 | 0.078↓ |
| 21813 | Tgfbr2 | 569.39 | 0.621 | 0.515↓ | 351.72 | 0.467 | 0.013↓* |
| 21808 | Tgfb2 | 71.23 | 0.128 | 0.012↓* | 93.69 | 0.166 | 0.019↓* |
| 18115 | Nnt | 45,948.78 | 2.120 | 0.001↑* | 37,695.71 | 1.660 | 0.008↑ |
| 23959 | Nt5e | 263.38 | 1.640 | 0.030↑ | 313.56 | 2.040 | 0.011↑* |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Gao, S.; Zhang, H.; Ren, Y.; Yu, X.; Lin, W.; Guo, S.; Zhu, R.; Wang, W. Decomposing the Mechanism of Qishen Granules in the Treatment of Heart Failure by a Quantitative Pathway Analysis Method. Molecules 2018, 23, 1829. https://doi.org/10.3390/molecules23071829
Ren W, Gao S, Zhang H, Ren Y, Yu X, Lin W, Guo S, Zhu R, Wang W. Decomposing the Mechanism of Qishen Granules in the Treatment of Heart Failure by a Quantitative Pathway Analysis Method. Molecules. 2018; 23(7):1829. https://doi.org/10.3390/molecules23071829
Chicago/Turabian StyleRen, Weiquan, Sheng Gao, Huimin Zhang, Yinglu Ren, Xue Yu, Weili Lin, Shuzhen Guo, Ruixin Zhu, and Wei Wang. 2018. "Decomposing the Mechanism of Qishen Granules in the Treatment of Heart Failure by a Quantitative Pathway Analysis Method" Molecules 23, no. 7: 1829. https://doi.org/10.3390/molecules23071829





