Solvent Extraction and Identification of Active Anticariogenic Metabolites in Piper cubeba L. through 1H-NMR-Based Metabolomics Approach
Abstract
:1. Introduction
2. Results
2.1. Yield of P. cubeba L. Extracts
2.2. Antibacterial Activity of P. cubeba L. Extracts
2.2.1. Disc Diffusion Assay
2.2.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.3. Liquid–Liquid Partition of P. cubeba L. Extract
2.4. Antibacterial Activity of P. cubeba L. Fractions
2.4.1. Disc Diffusion Assay
2.4.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
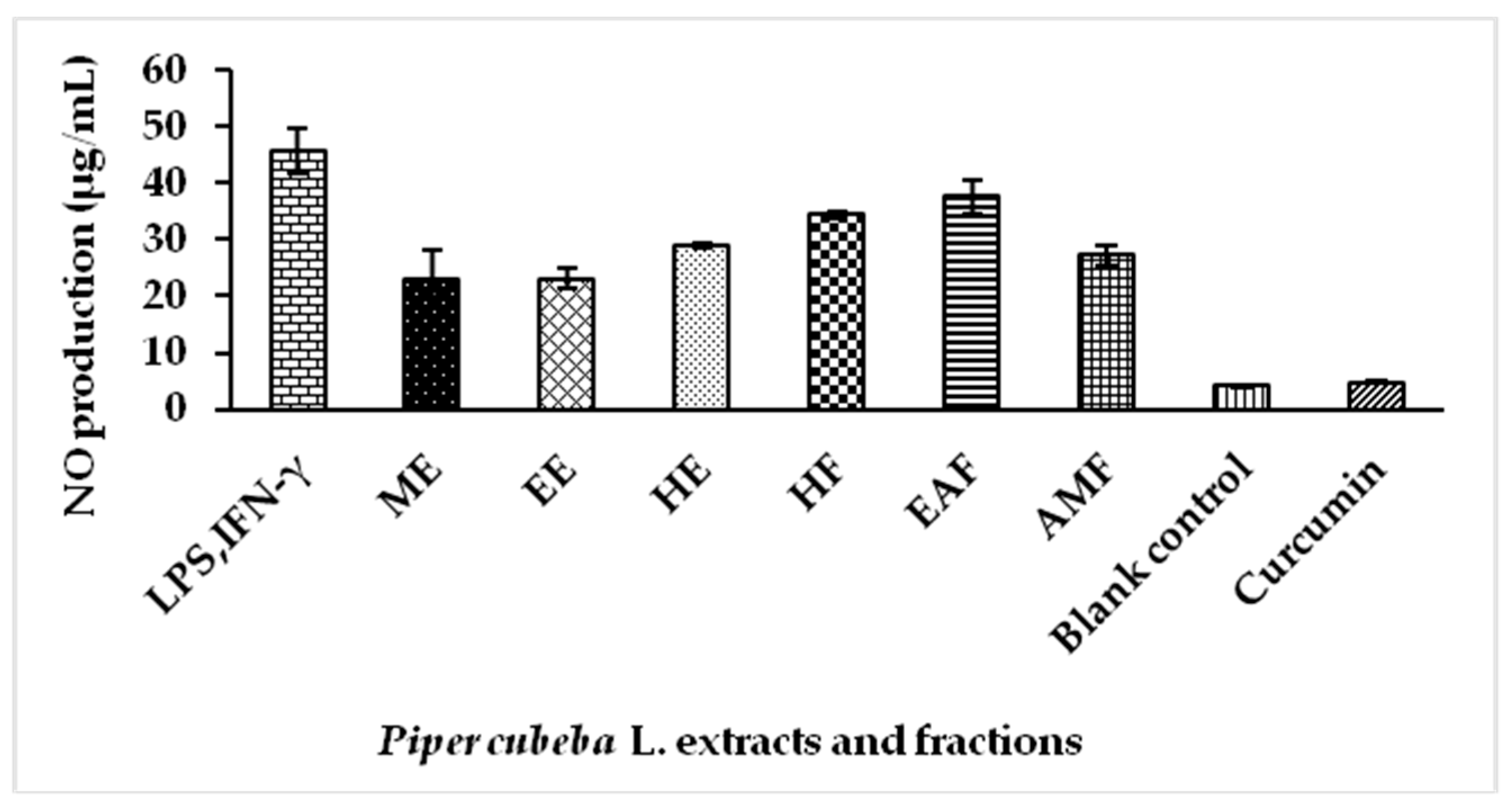
2.5. Nitric Oxide (NO) Production by Extracts and Fractions of P. cubeba L. in Lipopolysaccharide (LPS) Stimulated RAW 264.7 Cells
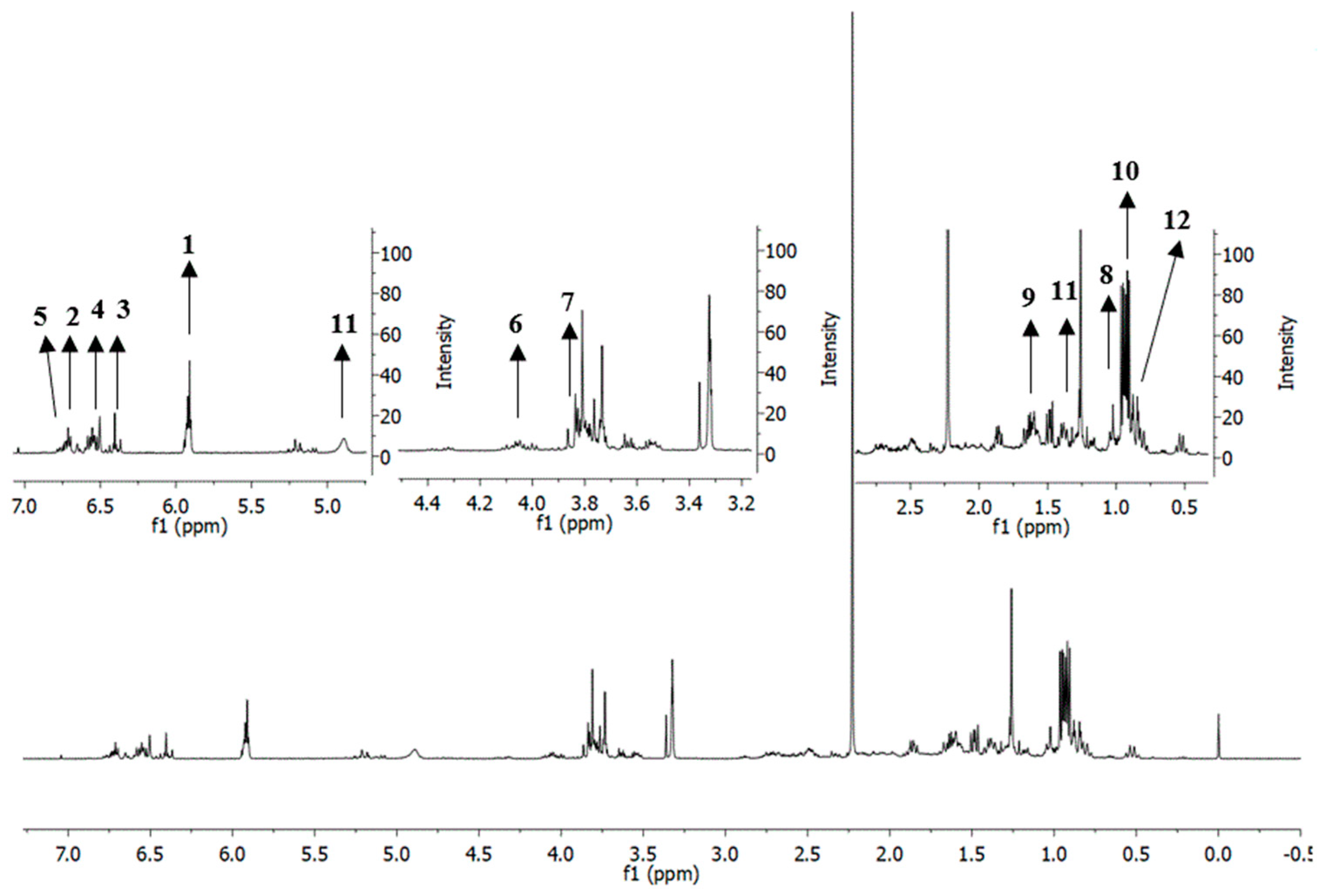
2.6. Putative Identification of Metabolites in Extracts and Fractions of P. cubeba L. Based on 1H-Nuclear Magnetic Resonance (NMR)
2.7. Multivariate Data Analysis (MVDA)
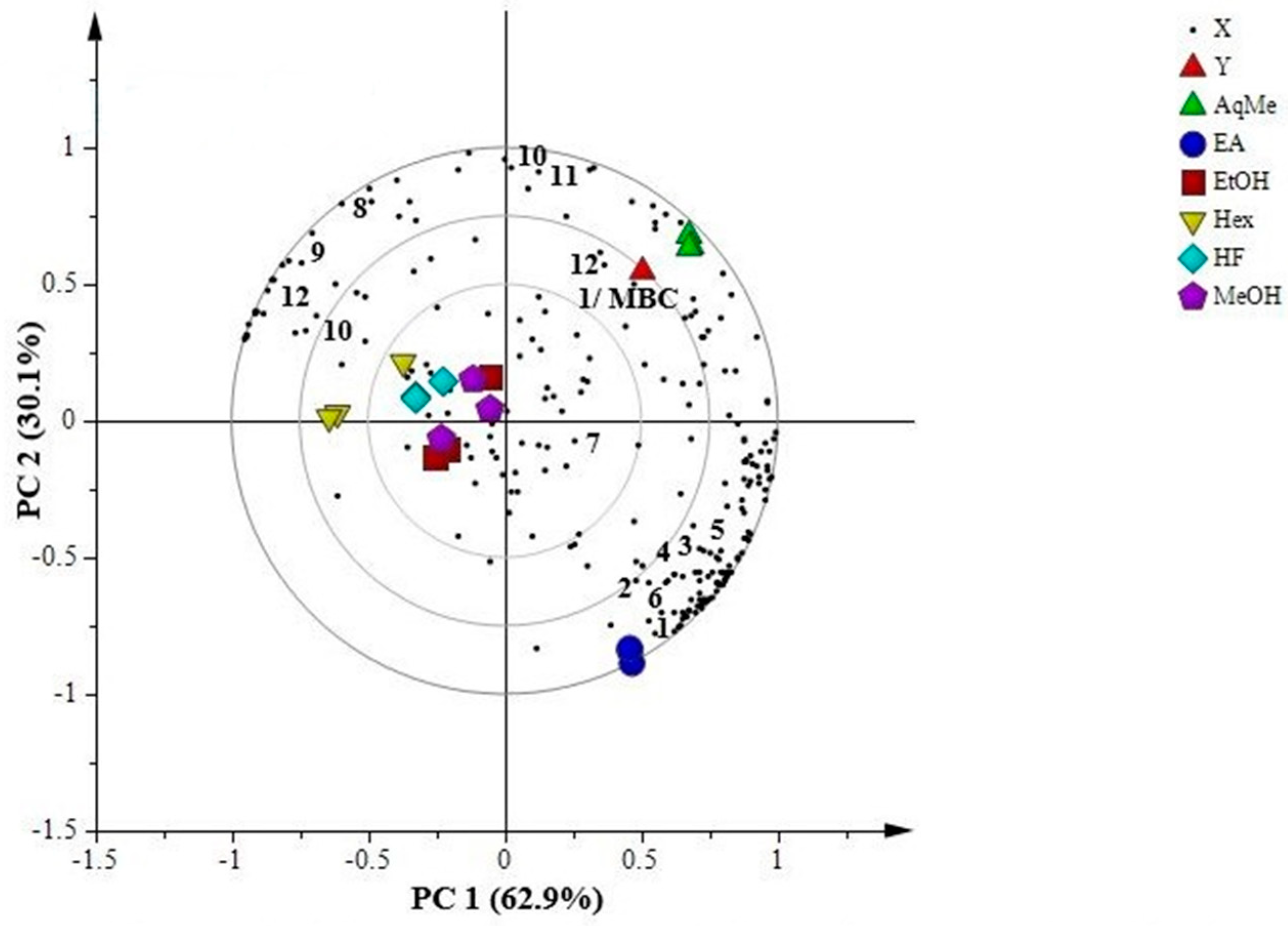
2.7.1. Principal Component Analysis (PCA)
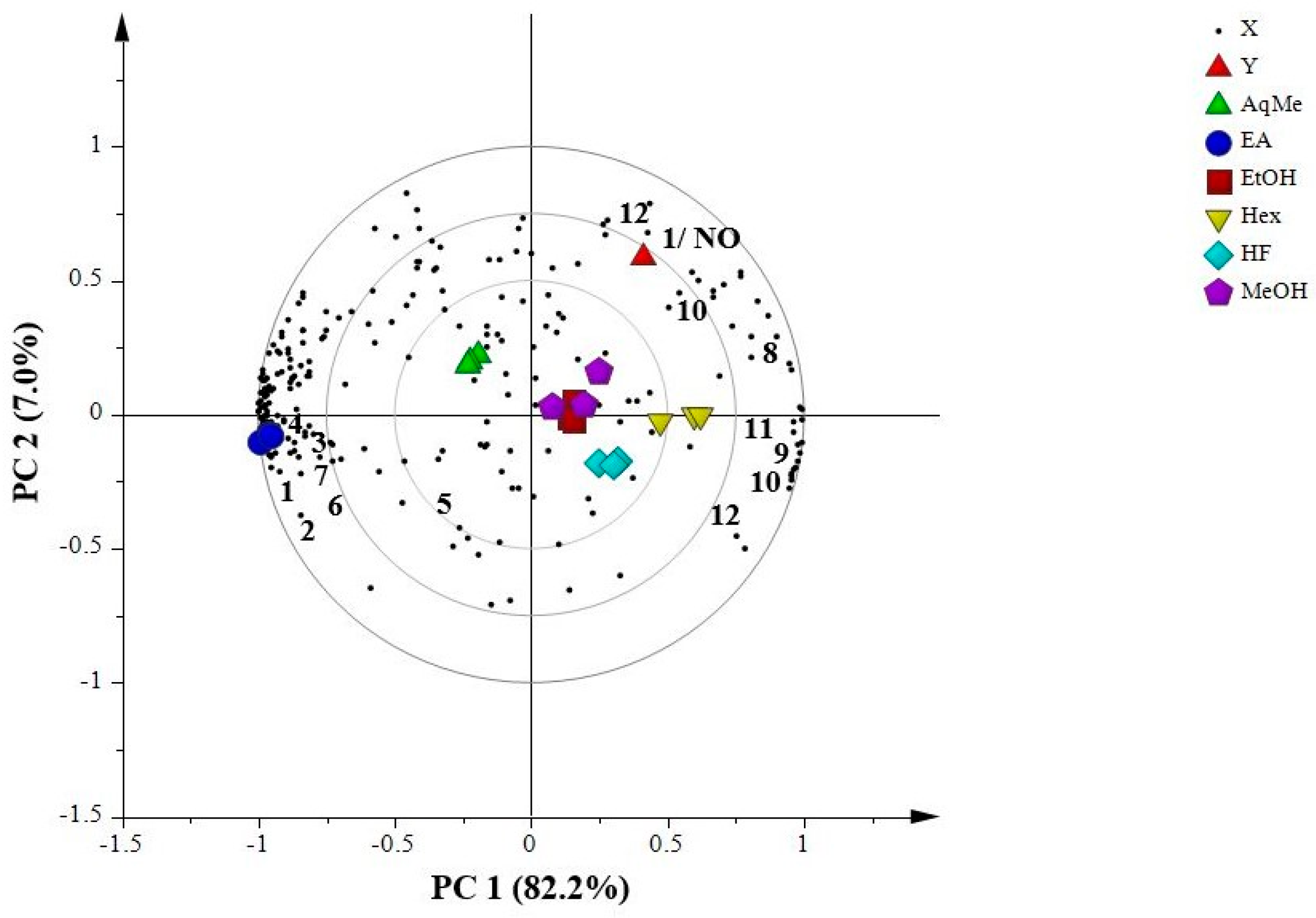
2.7.2. Partial Least Squares (PLS)
3. Materials and Methods
3.1. Materials
3.2. Plant Materials
3.3. Solvent Extraction of P. cubeba L.
3.4. Antibacterial Activity Assays
3.4.1. Sample Preparation
3.4.2. Disc Diffusion Assay
3.4.3. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
3.5. Liquid–Liquid Partition
3.6. Evaluation of Cytotoxic and Anti-Inflammatory Activity Using RAW 264.7 Murine Macrophage Cells Line
3.6.1. Sample Preparation
3.6.2. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
3.6.3. Nitric Oxide (NO) Production by Extracts and Fractions of P. cubeba L. Stimulated with LPS and IFN-γ
3.7. Proton Nuclear Magnetic Resonance (1H-NMR)
3.8. Multivariate Data Analysis (MVDA)
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forssten, S.D.; Bjorklund, M.; Ouwehand, A.C. Streptococcus mutans, caries and simulation models. Nutrients 2010, 2, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, T.M.; Szkaradkiewicz, A.K. Microbiology of dental caries. J. Biol. Earth Sci. 2013, 3, M21–M24. [Google Scholar]
- Bagramian, R.A.; Garcia-Godoy, F.; Volpe, A.R. The global increase in dental caries: A pending public health crisis. Am. J. Dent. 2009, 22, 3–8. [Google Scholar] [PubMed]
- Ambrosio, S.R.; Furtado, N.A.J.C.; De Oliveira, D.C.R.; Da Costa, F.B.; Martins, C.H.G.; De Carvalho, T.C.; Porto, T.S.; Veneziani, R.C.S. Antimicrobial activity of kaurane diterpenes against oral pathogens. J. Biosci. 2008, 63, 326–330. [Google Scholar] [CrossRef]
- More, G.; Tshikalange, T.E.; Lall, N.; Botha, F.; Meyer, J.J.M. Antimicrobial activity of medicinal plants against oral microorganisms. J. Ethnopharmacol. 2008, 119, 473–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porto, T.S.; Rangel, R.; Furtado, N.A.J.C.; De Carvalho, T.C.; Martins, C.H.G.; Veneziani, R.C.S.; Da Costa, F.B.; Vinholis, A.H.C.; Cunha, W.R.; Heleno, V.C.G.; et al. Pimarane-type diterpenes: Antimicrobial activity against oral pathogens. Molecules 2009, 14, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.; Dodds, M.; Tian, M. Naturally occurring phenolic antibacterial compounds show effectiveness against oral bacteria by a quantitative structure activity relationship study. J. Agric. Food Chem. 2008, 56, 11151–11156. [Google Scholar] [CrossRef] [PubMed]
- Yuliana, N.D.; Khatib, A.; Choi, Y.H.; Verpoorte, R. Metabolomics for bioactivity assessment of natural products. Phytother. Res. 2011, 25, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Rios, J.L.; Recio, M.C.; Villar, A. Screening methods for natural products with antimicrobial activity: A review of the literature. J. Ethnopharmacol. 1988, 23, 127–149. [Google Scholar] [CrossRef]
- Janssen, A.M.; Scheffer, J.J.C.; Baerheim, S.A. Antimicrobial activity of essential oils: A 1976–1986 literature review. Aspects of the test methods. Planta Medica 1987, 53, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Rukayadi, Y.; Lau, K.Y.; Zainin, N.S.; Zakaria, M.; Abas, F. Screening of antimicrobial activity of tropical edible medicinal plant extracts against five standard microorganisms for natural food preservatives. Int. Food Res. J. 2013, 20, 2905–2910. [Google Scholar]
- Saravia, M.E.; Nelson-Filho, P.; Ito, I.Y.; Da Silva, L.A.B.; Da Silva, R.A.B.; Emilson, C.G. Morphological differentiation between S. mutans and S. sobrinus on modified SB-20 culture medium. Microbiol. Res. 2011, 166, 63–67. [Google Scholar] [CrossRef] [PubMed]
- De Soet, J.J.; Toors, F.A.; De Graaff, J. Acidogenesis by oral streptococci at different pH values. Caries Res. 1989, 23, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Cohen, I.; Hay, D.I. Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect. Immun. 1986, 52, 555–561. [Google Scholar] [PubMed]
- Fang, L.; Weir, M.D.; Fouad, A.F.; Xu, H.H.K. Time-kill behaviour against eight bacterial species and cytotoxicity of antibacterial monomers. J. Dent. 2013, 41, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Chitnis, R.; Abichandani, M.; Nigam, P.; Nahar, L.; Sarker, S.D. Antioxidant and antibacterial activity of the extracts of Piper cubeba (Piperaceae). ARS Pharm. 2007, 48, 343–350. [Google Scholar]
- Addai, Z.R.; Abdullah, A.; Mutalib, S.A. Effect of extraction solvents on the phenolic content and antioxidant properties at two papaya cultivars. J. Med. Plants Res. 2013, 7, 3354–3359. [Google Scholar] [CrossRef]
- Graidist, P.; Martla, M.; Sukpondma, Y. Cytotoxic activity of Piper cubeba extract in breast cancer cell lines. Nutrients 2015, 7, 2707–2718. [Google Scholar] [CrossRef] [PubMed]
- Usia, T.; Iwata, H.; Hiratsuka, A.; Watabe, T.; Kadota, S.; Tezuka, Y. CYP3A4 and CYP2D6 inhibitory activities of Indonesian medicinal plants. Phytomedicine 2006, 13, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Marrtt, W.; Tan, G.T.; Gordell, G.A.; Pezzutuo, J.M. Biological activity of novel macrocylic alkaloids from Albizia amara detected on the basis of interaction with DNA. J. Nat. Prod. 1991, 54, 1531–1542. [Google Scholar] [CrossRef]
- Farag, R.S.; Daw, Z.Y.; Hewed, F.M.; Elbaraty, G.S. Antimicrobial activity of some Egyptian spice essential oil. J. Food Prot. 1989, 52, 665–667. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [PubMed]
- Rintu, D.; Shinjini, M.; Kaustab, M.; Pramathadhip, P.; Umesh, P.S.; Banerjee, E.R. Anti-oxidant and anti-inflammatory activities of different varieties of Piper leaf extracts (Piper betle L.). J. Nutr. Food Sci. 2015, 5, 415. [Google Scholar] [CrossRef]
- Tewtrakul, S.; Itharat, A. Nitric oxide inhibitory substances from the rhizomes of Dioscorea membranacea. J. Ethnopharmacol. 2007, 109, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Kang, T.H.; Song, E.K.; Pae, H.O.; Chung, H.T.; Kim, Y.C. Inhibitory effects of butanol fraction of the aqueous extract of Forsythia koreana on the nitric oxide production by murine macrophage-like RAW 264.7 cells. J. Ethnopharmacol. 2000, 73, 323–327. [Google Scholar] [CrossRef]
- Conforti, F.; Menichini, F. Phenolic compounds from plants as nitric oxide production inhibitors. Curr. Med. Chem. 2011, 18, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Robbins, K.S.; Greenspan, P.; Pegg, R.B. Effect of pecan phenolics on the release of nitric oxide from murine RAW 264.7 macrophage cells. Food Chem. 2016, 212, 681–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delehanty, J.B.; Johnson, B.J.; Hickey, T.E.; Pons, T.; Ligler, F.S. Binding and neutralization of lipopolysaccharides by plant proanthocyanidins. J. Nat. Prod. 2007, 70, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Terra, X.; Valls, J.; Vitrac, X.; Merrillon, J.M.; Arola, L.; Ardevol, A. Grape-seed procyanidins act as antiinflammatory agents in endotoxinstimulated RAW 264.7 macrophages by inhibiting NFeB signaling pathway. J. Agric. Food Chem. 2007, 55, 4357–4365. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Rimbach, G.; Saliou, C.; Valacchi, G.; Packer, L. Activity of monomeric, dimeric, and trimeric flavonoids on NO production, TNF-alpha secretion, and NF-kappaB-dependent gene expression in RAW 264.7 macrophages. FEBS Lett. 2000, 465, 93–97. [Google Scholar] [CrossRef]
- Silverstein, R.M.; Webster, F.X.; Kiemle, D.J. Proton NMR Spectrometry. In Spectrometric Identification of Organic Compounds, 7th ed.; John Wiley & Sons Inc.: New York, NY, USA, 2005; p. 142. ISBN 0-471-39362-2. [Google Scholar]
- Mulik, M.B.; Laddha, K.S. Isolation, characterization and quantification of bioactive dibenzylbutyrolactone lignan (−)-cubebin from fruits of Piper cubeba L.f. Indian J. Nat. Prod. Resour. 2015, 6, 189–193. [Google Scholar]
- Ruslan, K.; Batterman, S.; Bos, R.; Kayser, O.; Woerdenbag, H.J.; Quax, W.J. Lignan profile of Piper cubeba, an Indonesian medicinal plant. Biochem. Syst. Ecol. 2007, 35, 397–402. [Google Scholar] [CrossRef]
- Junqueira, A.P.F.; Perazzo, F.F.; Souza, G.H.B.; Maistro, E.L. Clastogenicity of Piper cubeba (Piperaceae) seed extract in an in vivo mammalian cell system. Genet. Mol. Biol. 2007, 30, 656–663. [Google Scholar] [CrossRef]
- Parmar, V.S.; Jain, S.C.; Bisht, K.S.; Jain, R.; Taneja, P.; Jha, A. Phytochemistry of the genus Piper. Phytochemistry 1997, 46, 597–673. [Google Scholar] [CrossRef]
- Bos, R.; Woerdenbag, H.J.; Kayser, O.; Quax, W.J.; Ruslan, K.; Elfahmi. Essential oil constituents of Piper cubeba L. fils. from Indonesia. J. Essent. Oil Res. 2007, 19, 14–17. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Maulidiani, M.; Abas, F.; Khatib, A.; Shitan, M.; Shaari, K.; Lajis, N.H. Comparison of partial least squares and artificial neural network for the prediction of antioxidant activity in extract of pegaga (Centella) varieties from 1H-Nuclear Magnetic Resonance spectroscopy. Food Res. Int. 2013, 54, 852–860. [Google Scholar] [CrossRef]
- Choi, H.; Choi, Y.H.; Verberne, M.; Lefeber, A.W.M.; Erkelens, C.; Verpoorte, R. Metabolic fingerprinting of wild type and transgenic tobacco plants by 1H-NMR and multivariate analysis technique. Phytochemistry 2004, 65, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Mediani, A.; Abas, F.; Khatib, A.; Tan, C.P.; Ismail, I.S.; Shaari, K.; Lajis, N.H. Phytochemical and biological features of Phyllanthus niruri and Phyllanthus urinaria harvested at different growth stages revealed by 1H-NMR-based metabolomics. Ind. Crops Prod. 2015, 77, 602–613. [Google Scholar] [CrossRef]
- Fonville, J.M.; Richards, S.E.; Barton, R.H.; Boulange, C.L.; Ebbels, T.M.D.; Nicholson, J.K. The evolution of partial least squares models and related chemometric approaches in metabonomics and metabolic phenotyping. J. Chemom. 2010, 24, 636–649. [Google Scholar] [CrossRef]
- Andrade, B.F.M.T.; Nunes Barbosa, L.; Da Silva Probst, I.; Fernandes Junior, A. Antimicrobial activity of essential oils. J. Essent. Oil Res. 2014, 26, 34–40. [Google Scholar] [CrossRef]
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Idaomar, M. Biological effects of essential oils—A review. Food Chem. Toxicol. 2008, 46, 446–475. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.K.; Akhtar, M.S.; Sinniah, U.R. Antimicrobial properties of plant essential oils against human pathogens and their mode of action: An updated review. Evid. Based Complement. Altern. Med. 2016, 2016, 3012462. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef] [PubMed]
- Dorman, H.J.; Deans, S.G. Antimicrobial agents from plants: Antibacterial activity of plant volatile oils. J. Appl. Microbiol. 2000, 88, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.; Cabral, C.; Evora, L.; Ferreira, I.; Cavaleiro, C.; Cruz, M.T. Chemical composition, anti-inflammatory activity and cytotoxicity of Thymus zygis L. subsp. Sylvestris (Hoffmanns. & Link) Cout. essential oil and its main compounds. Arab. J. Chem. 2015. [Google Scholar] [CrossRef]
- Bonjardim, L.R.; Cunha, E.S.; Guimareas, A.G.; Santana, M.F.; Oliveira, M.G.B.; Serafini, M.R. Evaluation of the anti-inflammatory and antinociceptive properties of p-cymene in mice. Z. Naturforsch. 2012, 15–20. [Google Scholar] [CrossRef]
- Lee, H.J.; Jeong, H.S.; Kim, D.J.; Noh, Y.H.; Yuk, D.Y.; Hong, J.T. Inhibitory effect of citral on NO production by suppression of iNOS expression and NF-κB activation in RAW 264.7 cells. Arch. Pharm. Res. 2008, 31, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Mothana, R.A.; Al-Said, M.S.; Raish, M.; Khaled, J.M.; Alharbi, N.S.; Alatar, A. Chemical composition, anti-inflammatory and antioxidant activities of the essential oil of Piper cubeba L. Rom. Biotechnol. Lett. 2017, 22, 12366–12376. [Google Scholar]
- Dona, A.C.; Kyriakides, M.; Scott, F.; Shephard, E.A.; Varshavi, D.; Veselkov, K.; Everett, J.R. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput. Struct. Biotechnol. J. 2016, 14, 135–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maulidiani, M.; Sheikh, B.Y.; Mediani, A.; Wei, L.S.; Ismail, I.S.; Abas, F.; Lajis, N.H. Differentiation of Nigella sativa seeds from four different origins and their bioactivity correlations based on NMR-metabolomics approach. Phytochem. Lett. 2015, 13, 308–318. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikstrom, C.; Wold, S. Multi and Megavariate Data Analysis Part I: Basic Principles and Applications, 3rd ed.; Umetrics Inc.: Malmo, Sweden, 2006; p. 425. ISBN 9197373028. [Google Scholar]
- Rukayadi, Y.; Shim, J.S.; Hwang, J.K. Screening of Thai medicinal plants for anticandidal activity. Mycoses 2008, 51, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard-Ninth Edition; CLSI Document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard-Eleventh Edition; CLSI Document M02-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- American Type Culture Collection (ATCC). ATCC Animal Cell Culture Guide; American Type Culture Collection: Manassas, VA, USA, 2014. [Google Scholar]
- Tan, W.S.; Arulselvan, P.; Karthivashan, G.; Fakurazi, S. Moringa oleifera flower extract suppresses the activation of inflammatory mediators in lipopolysaccharide-stimulated RAW 264.7 macrophages via NF-κB pathway. Mediat. Inflamm. 2015, 2015, 720171. [Google Scholar] [CrossRef] [PubMed]
- Abas, F.; Lajis, N.H.; Israf, D.A.; Shaari, K.; Umi Kalsom, Y. Antioxidant and nitric oxide inhibition activities of selected Malay traditional vegetables. Food Chem. 2006, 95, 566–573. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Khatib, A.; Maulidiani, H.; Shaari, K.; Choi, Y.H.; Lajis, N.H. 1H-NMR-based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res. Int. 2012, 49, 763–770. [Google Scholar] [CrossRef]
- Souza, G.H.B.; Da Silva Filho, A.A.; De Souza, V.A.; Pereira, A.C.; Royo, V.D.A.; Silva, M.L.A. Analgesic and anti-inflammatory activities evaluation of (−)-O-acetyl, (−)-O-methyl, (−)-O-dimethylethylamine cubebin and their preparation from (−)-cubebin. IL Farmaco 2004, 59, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Itoh, K.; Tachibana, S. Extractives of Juniperus chinensis L. I: Isolation of podophyllotoxin and yatein from the leaves of J. chinensis. J. Wood Sci. 1998, 44, 397–400. [Google Scholar] [CrossRef]
- De Souza, V.A.; Da Silva, R.; Pereira, A.C.; Royo, V.D.A.; Saraiva, J.; Montanheiro, M. Trypanocidal activity of (−)-cubebin derivatives against free amastigote forms of Trypanosoma cruzi. Bioorg. Med. Chem. Lett. 2005, 15, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Laurentiz, R.S.; Borges, A.; Laurentiz, A.C.; Silva, M.L.A.; Martins, C.H.G. Evaluation of antimycobacterial activity of dihydrocubebin lignan extracted from Piper cubeba and its semisynthetic derivatives. Rev. Bras. Plantas Med. 2015, 17, 782–789. [Google Scholar] [CrossRef]
- Prabhu, B.R.; Mulchandani, N.B. Lignans from Piper cubeba. Phytochemistry 1985, 24, 329–331. [Google Scholar] [CrossRef]
- Ryu, J.H.; Ja Son, H.; Lee, S.H.; Sohn, D.H. Two neolignans from Perilla frutescens and their inhibition of nitric oxide synthase and tumor necrosis factor-α expression in murine macrophage cell line RAW 264.7. Bioorg. Med. Chem. Lett. 2002, 12, 649–651. [Google Scholar] [CrossRef]
- Human Metabolome Database (HMDB). HMDB 05805. Available online: http://www.hmdb.ca/metabolites/HMDB05805 (accessed on 29 November 2017).
- Human Metabolome Database (HMDB). HMDB 0034301. Available online: http://www.hmdb.ca/metabolites/HMDB34301 (accessed on 29 November 2017).
- Chen, X.H.; Kim, C.S.; Kashiwagi, T.; Tebayashi, S.I.; Horiike, M. Antifeedants against Acusta despesta from the Japanese cedar, Cryptomeria japonica II. Biosci. Biotechnol. Biochem. 2001, 65, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Kentaro, O.; Shimazaki, K.; Chuman, T. X-ray crystallography and NOE studies on the conformation of periplanones and their analogues. J. Chem. Soc. Perkin Trans. 1 1990, 1769–1777. [Google Scholar] [CrossRef]
- Miyazawa, M.; Uemura, T.; Kameoka, H. Biotransformation of sesquiterpenoids (−)-globulol and (+)-ledol by Glomerella cingulate. Phytochemistry 1994, 37, 1027–1030. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| Dried Berries (g) | Solvent | Extraction Step | Yield (g) | Yield (%) |
|---|---|---|---|---|
| 100 | Methanol | First extraction | 18.84 | 18.84 |
| 750 | Methanol | Second extraction | 164.46 | 21.93 |
| 100 | Ethanol | First extraction | 15.71 | 15.71 |
| 100 | Hexane | First extraction | 9.91 | 9.91 |
| Inhibition Zone (mm) ± SD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Conc. (mg/mL) | Streptococcus mutans KCCM 3390 | Streptococcus sobrinus ATCC33478 | Actinomyces viscosus ATCC15987 | ||||||
| ME | EE | HE | ME | EE | HE | ME | EE | HE | |
| 6.3 | 9.33 ± 0.58 AB a | 10.00 ± 0.00 AB a | 8.33 ± 0.29 B b | 7.00 ± 0.00 C a | 7.00 ± 0.00 D a | 7.10 ± 0.17 C a | 9.00 ± 0.00 C a | 9.00 ± 0.00 C a | 9.67 ± 0.58 C a |
| 12.5 | 9.67 ± 0.58 AB a | 9.33 ± 0.58 AB a | 10.00 ± 0.00 A a | 9.00 ± 0.00 B a | 7.00 ± 0.00 D c | 8.10 ± 0.10 B b | 10.17 ± 0.29 BC a | 8.83 ± 0.76 C a | 9.67 ± 0.58 C a |
| 25.0 | 9.00 ± 0.00 B b | 9.17 ± 0.29 B b | 10.00 ± 0.00 A a | 9.00 ± 0.00 B a | 8.67 ± 0.58 C a | 8.43 ± 0.40 B a | 10.67 ± 0.58 AB a | 10.00 ± 0.00 BC a | 10.50 ± 0.50 BC a |
| 50.0 | 10.00 ± 0.00 A a | 10.00 ± 0.00 AB a | 10.00 ± 0.00 A a | 11.67 ± 0.58 A a | 10.00 ± 0.00 B b | 12.03 ± 0.06 A a | 11.50 ± 0.50 A a | 11.33 ± 0.58 A a | 11.33 ± 0.58 AB a |
| 100 | 10.00 ± 0.00 A a | 10.33 ± 0.58 A a* | 10.00 ± 0.00 A a | 12.17 ± 0.06 A a | 11.13 ± 0.15 A b | 12.07 ± 0.12 A a | 11.67 ± 0.58 A ab | 11.17 ± 0.29 AB b | 12.67 ± 0.58 A a* |
| CHX (0.05 mg/mL) | 11.00 ± 0.00 * | 11.00 ± 0.00 * | 11.00 ± 0.00 * | 14.00 ± 0.00 * | 14.00 ± 0.00 * | 14.00 ± 0.00 * | 13.33 ± 0.58 * | 13.33 ± 0.58 * | 13.33 ± 0.58 * |
| Bacteria | ME | EE | HE | CHX (µg/mL) | |
|---|---|---|---|---|---|
| S. mutans KCCM3309 | MIC (mg/mL) MBC (mg/mL) | 0.23 ± 0.15 0.23 ± 0.15 | 0.10 ± 0.00 0.10 ± 0.00 | 0.10 ± 0.00 0.10 ± 0.00 | 1.14 ± 0.75 1.14 ± 0.75 |
| S. sobrinus ATCC33478 | MIC (mg/mL) MBC (mg/mL) | 0.93 ± 0.61 16.67 ± 7.22 | 0.80 ± 0.69 20.83 ± 7.22 | 0.87 ± 0.70 20.83 ± 7.22 | 0.49 ± 0.00 2.77 ± 1.97 |
| A. viscosus ATCC15987 | MIC (mg/mL) MBC (mg/mL) | 0.80 ± 0.69 20.83 ± 7.22 | 0.47 ± 0.31 20.83 ± 7.22 | 0.87 ± 0.70 13.54 ± 10.97 | 1.14 ± 0.75 1.63 ± 0.57 |
| Methanol Extract (g) | Solvent | Yield (g) | Yield (%) |
|---|---|---|---|
| 120 | Hexane | 46.56 | 35.82 |
| Ethyl acetate | 13.96 | 10.74 | |
| Aqueous methanol | 2.33 | 2.59 |
| Inhibition Zone (mm) ± SD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Conc. (mg/mL) | Streptococcus mutans KCCM 3390 | Streptococcus sobrinus ATCC33478 | Actinomyces viscosus ATCC15987 | ||||||
| HF | EAF | AMF | HF | EAF | AMF | HF | EAF | AMF | |
| 6.3 | 7.00 ± 0.00 B a | 7.00 ± 0.00 B a | 7.00 ± 0.00 D a | 7.10 ± 0.17 C a | 7.07 ± 0.06 D a | 7.00 ± 0.00 C a | 7.00 ± 0.00 C a | 7.00 ± 0.00 C a | 7.00 ± 0.00 D a |
| 12.5 | 9.33 ± 0.58 A a | 9.17 ± 0.29 A a | 9.33 ± 0.58 C a | 7.13 ± 0.06 C a | 7.10 ± 0.10 CD a | 7.10 ± 0.10 C a | 7.67 ± 0.58 C b | 9.33 ± 0.58 B a | 9.33 ± 0.58 C a |
| 25.0 | 9.00 ± 0.00 A b | 10.00 ± 0.00 A a | 9.83 ± 0.29 BC a | 7.33 ± 0.58 C b | 7.27 ± 0.06 C b | 8.67 ± 0.58 B a | 9.33 ± 0.58 B b | 10.33 ± 0.58 AB ab | 11.00 ± 0.00 B a |
| 50.0 | 9.00 ± 0.00 A b | 9.83 ± 0.29 A ab | 10.50 ± 0.50 AB a* | 10.00 ± 0.00 B a | 10.00 ± 0.00 B a | 10.67 ± 0.58 A a | 10.00 ± 0.00 B b | 10.33 ± 0.58 AB ab | 11.17 ± 0.29 B a |
| 100 | 9.33 ± 0.58 A b | 9.66 ± 0.58 A b | 11.00 ± 0.00 A a* | 11.13 ± 0.12 A a | 11.07 ± 0.06 A a | 11.20 ± 0.17 A a | 12.00 ± 0.00 A a | 10.83 ± 0.29 A b | 12.33 ± 0.58 A a* |
| CHX (0.05 mg/mL) | 11.00 ± 0.00 * | 11.00 ± 0.00 * | 11.00 ± 0.00 * | 14.00 ± 0.00 * | 14.00 ± 0.00 * | 14.00 ± 0.00 * | 13.33 ± 0.58 * | 13.33 ± 0.58 * | 13.33 ± 0.58 * |
| Bacteria | HF | EAF | AMF | CHX (µg/mL) | |
|---|---|---|---|---|---|
| S. mutans KCCM3309 | MIC (mg/mL) MBC (mg/mL) | 0.10 ± 0.00 0.10 ± 0.00 | 0.13 ± 0.06 0.13 ± 0.06 | 0.10 ± 0.00 0.10 ± 0.00 | 1.14 ± 0.75 1.14 ± 0.75 |
| S. sobrinus ATCC33478 | MIC (mg/mL) MBC (mg/mL) | 0.67 ± 0.23 25.00 ± 0.00 | 3.15 ± 2.68 25.00 ± 0.00 | 1.07 ± 0.46 7.29 ± 4.77 | 0.49 ± 0.00 2.77 ± 1.97 |
| A. viscosus ATCC15987 | MIC (mg/mL) MBC (mg/mL) | 1.58 ± 1.35 16.67 ± 7.22 | 1.20 ± 0.69 25.00 ± 0.00 | 1.07 ± 0.46 12.50 ± 10.83 | 1.14 ± 0.75 1.63 ± 0.57 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raja Mazlan, R.N.A.; Rukayadi, Y.; Maulidiani, M.; Ismail, I.S. Solvent Extraction and Identification of Active Anticariogenic Metabolites in Piper cubeba L. through 1H-NMR-Based Metabolomics Approach. Molecules 2018, 23, 1730. https://doi.org/10.3390/molecules23071730
Raja Mazlan RNA, Rukayadi Y, Maulidiani M, Ismail IS. Solvent Extraction and Identification of Active Anticariogenic Metabolites in Piper cubeba L. through 1H-NMR-Based Metabolomics Approach. Molecules. 2018; 23(7):1730. https://doi.org/10.3390/molecules23071730
Chicago/Turabian StyleRaja Mazlan, Raja Nur Asila, Yaya Rukayadi, M. Maulidiani, and Intan Safinar Ismail. 2018. "Solvent Extraction and Identification of Active Anticariogenic Metabolites in Piper cubeba L. through 1H-NMR-Based Metabolomics Approach" Molecules 23, no. 7: 1730. https://doi.org/10.3390/molecules23071730





