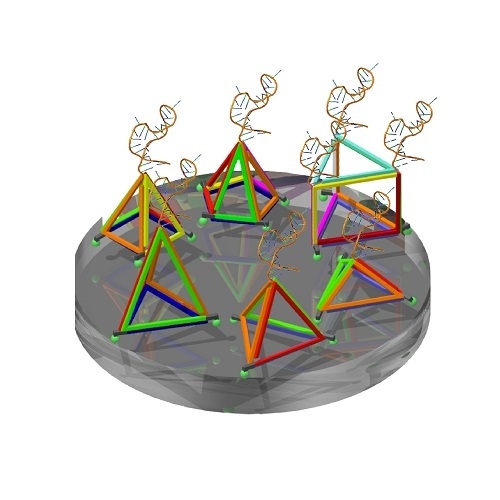
Aptamer Display on Diverse DNA Polyhedron Supports
Abstract
:1. Introduction
2. Results
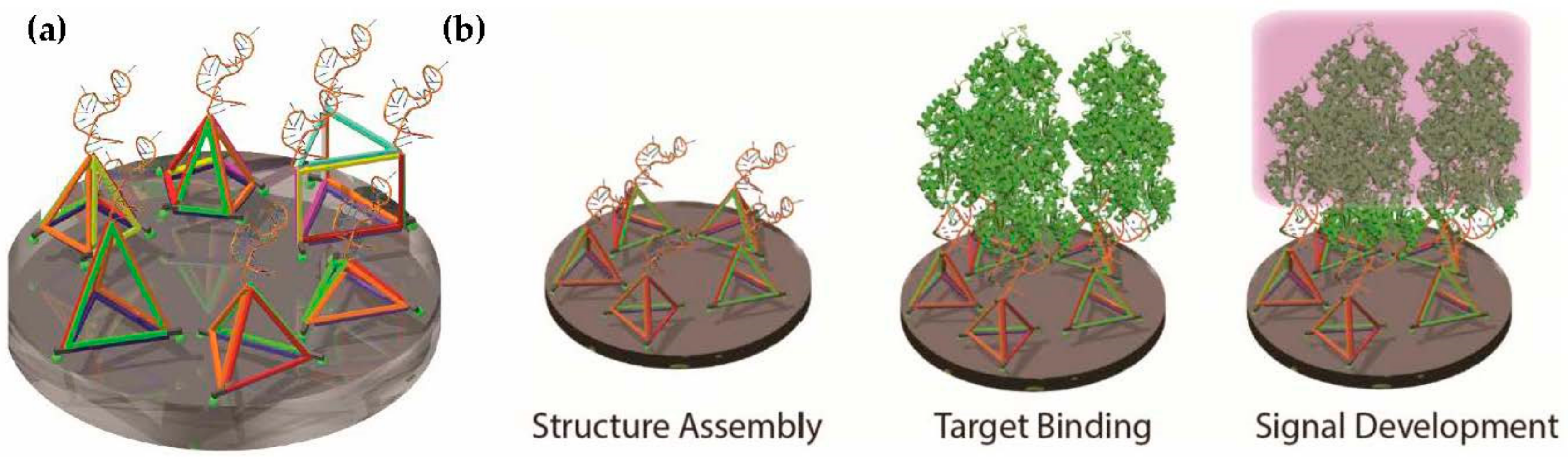
2.1. Design and Assembly of Aptamer-Integrated DNA Nanostructures
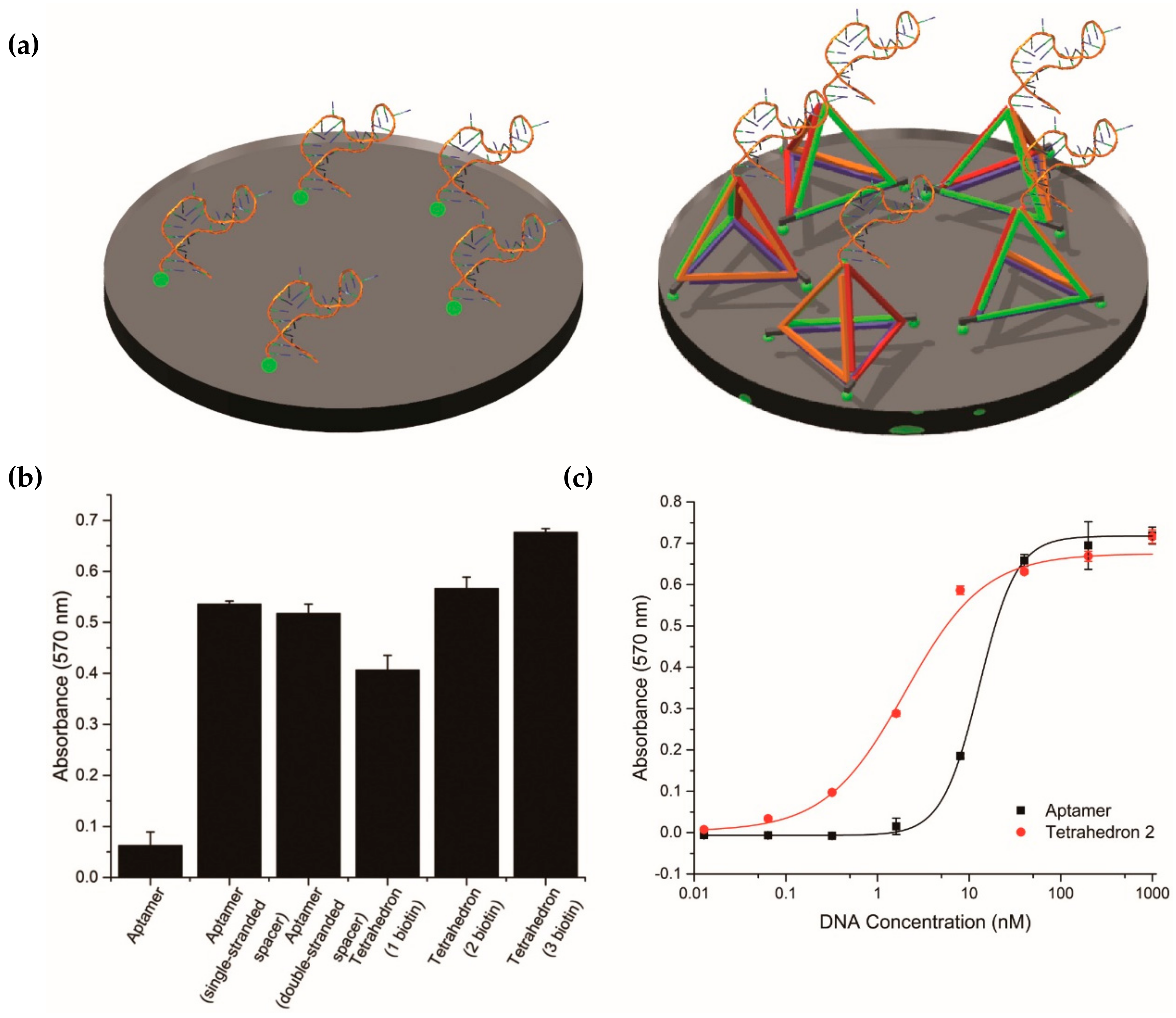
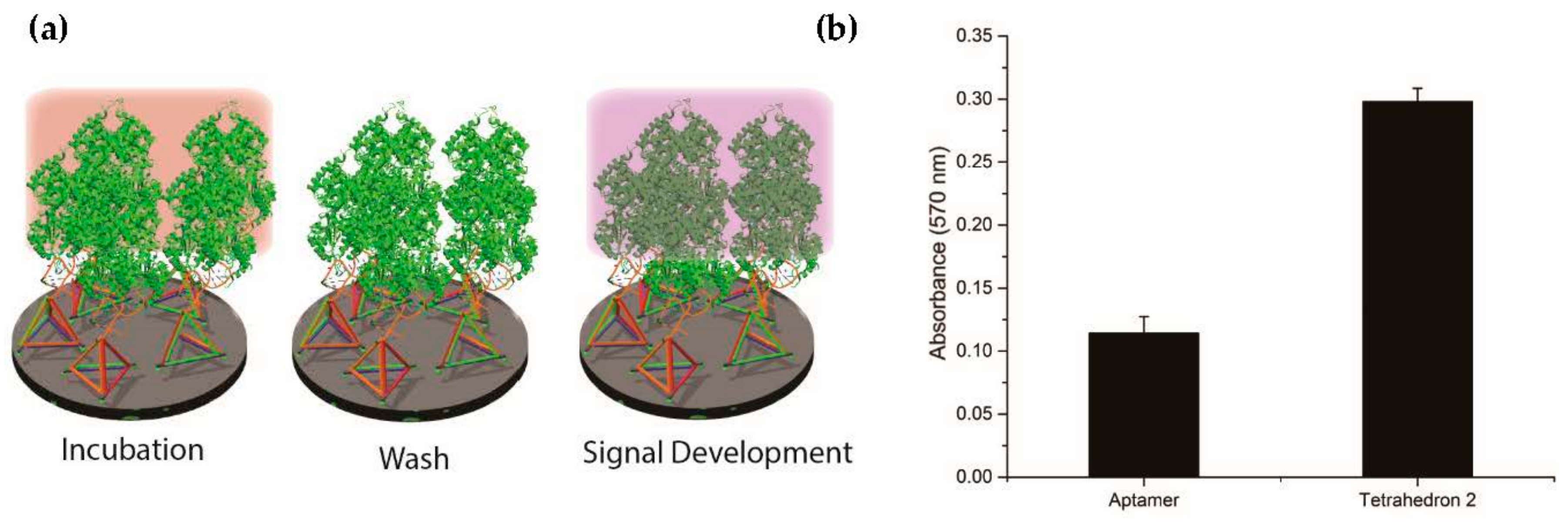
2.2. Position of Aptamer-Tetrahedron on Surface and Comparison with Single-Stranded Aptamer
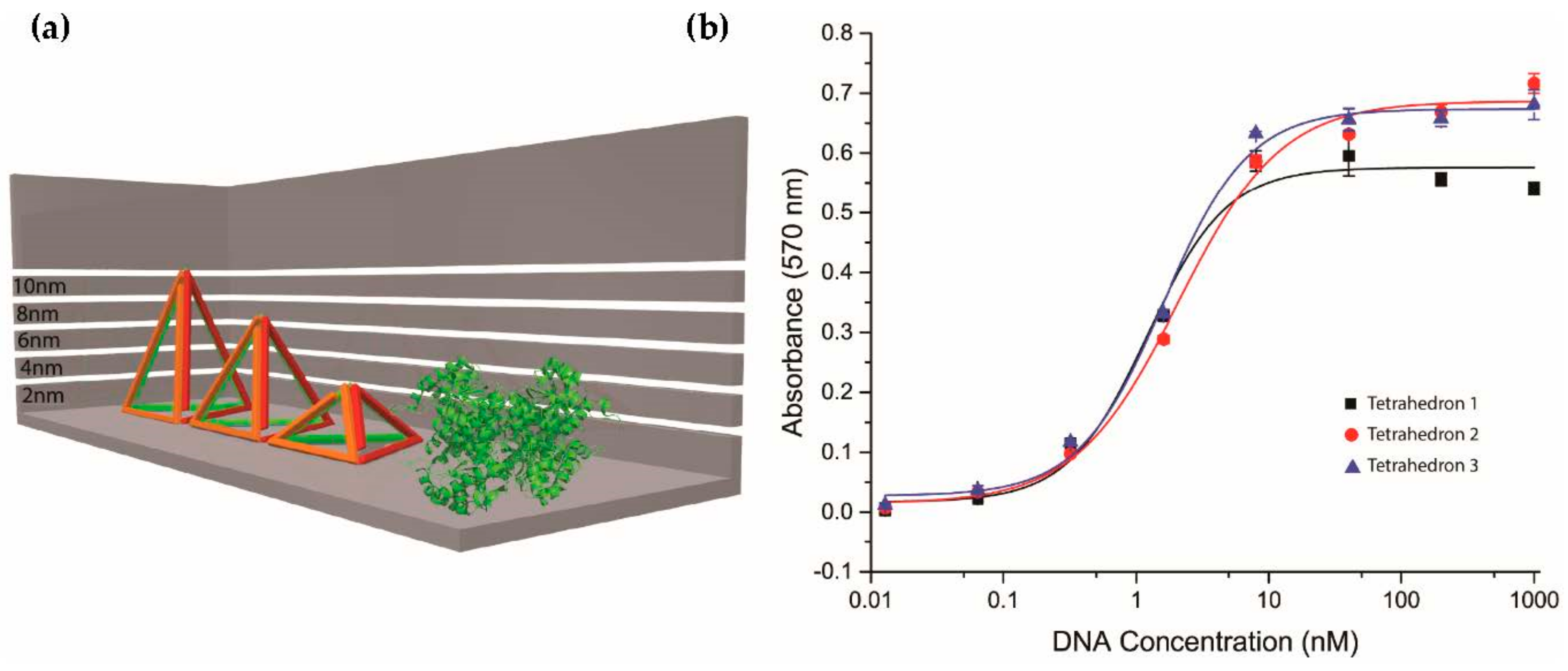
2.3. Investigation of Different Heights of Tetrahedron
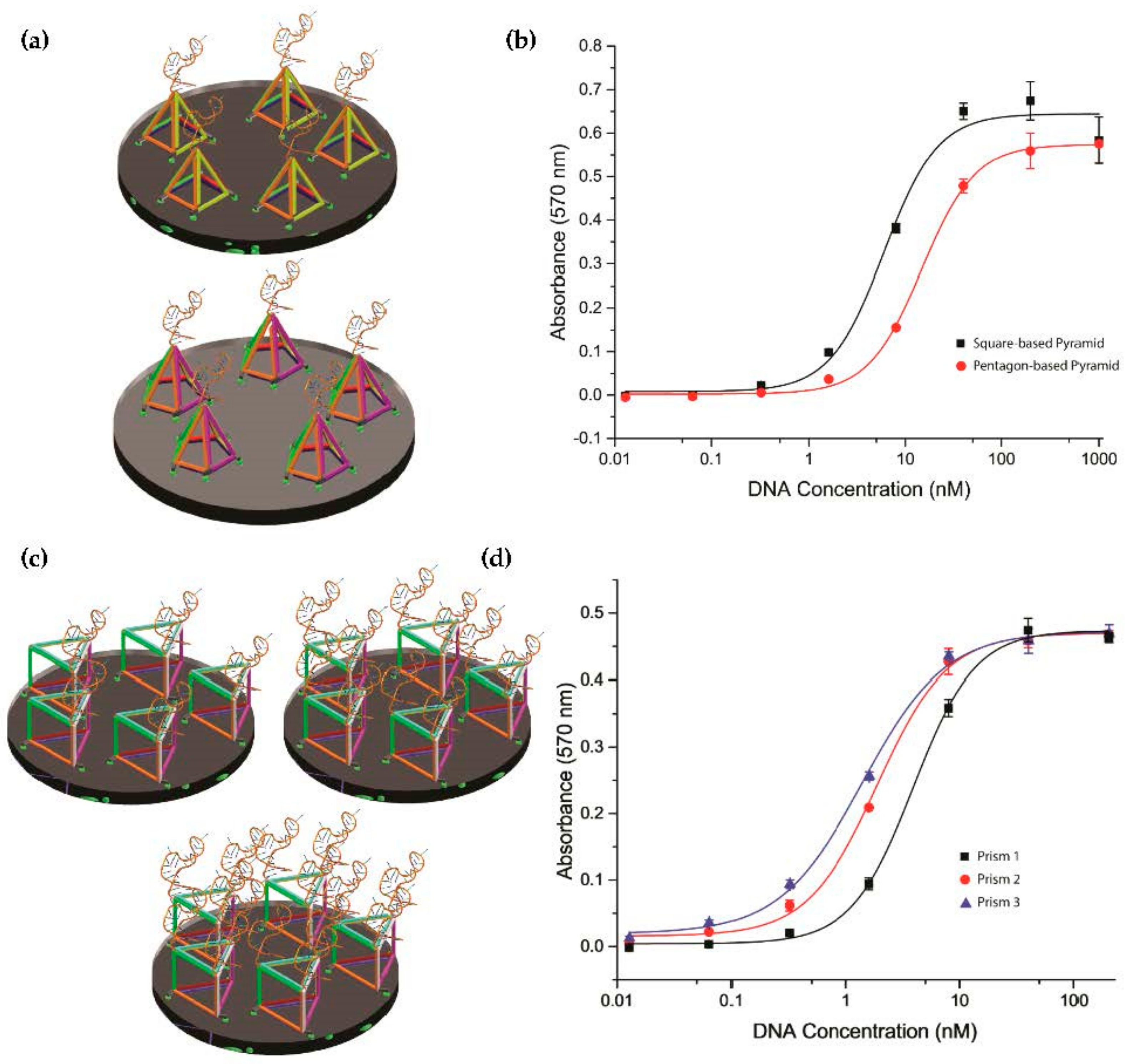
2.4. Investigation of Tetrahedron Compared to Other Three-Dimensional Nanostructure Polyhedra
2.5. Detection of Exogenous Recombinant PfLDH in Whole Rat Blood
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides, Enzymes and Other Chemicals
4.2. Assembly of DNA Nanostructures
4.3. Polyacrylamide Gel Electrophoresis (PAGE) Showing the Formation of DNA Nanostructures
4.4. Aptamer-DNA Nanostructure Functionalization of 96-Well Plates
4.5. APTEC Assay Protocol
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Frosch, A.E.P.; John, C.C. Immunomodulation in Plasmodium falciparum malaria: Experiments in nature and their conflicting implications for potential therapeutic agents. Expert Rev. Anti-Infect. Ther. 2012, 10, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Preuss, J.; Jortzik, E.; Becker, K. Glucose-6-phosphate metabolism in Plasmodium falciparum. IUBMB Life 2012, 64, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.W.; Kwok, J.; Law, A.W.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Dirkzwager, R.M.; Kinghorn, A.B.; Richards, J.S.; Tanner, J.A. APTEC: Aptamer-tethered enzyme capture as a novel rapid diagnostic test for malaria. Chem. Commun. 2015, 51, 4697–4700. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.-W.; Dirkzwager, R.M.; Wong, W.-C.; Cardoso, J.; D’Arc Neves Costa, J.; Tanner, J.A. Aptamer-mediated Plasmodium-specific diagnosis of malaria. Biochimie 2018, 145, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Miranda, G.; Feng, L.Y.; Shiu, S.C.C.; Dirkzwager, R.M.; Cheung, Y.W.; Tanner, J.A.; Schoning, M.J.; Offenhausser, A.; Mayer, D. Aptamer-based electrochemical biosensor for highly sensitive and selective malaria detection with adjustable dynamic response range and reusability. Sens. Actuator B Chem. 2018, 255, 235–243. [Google Scholar] [CrossRef]
- Shiu, S.C.C.; Cheung, Y.W.; Dirkzwager, R.M.; Liang, S.; Kinghorn, A.B.; Fraser, L.A.; Tang, M.S.L.; Tanner, J.A. Aptamer-mediated protein molecular recognition driving a DNA tweezer nanomachine. Adv. Biosyst. 2017, 1, 1600006. [Google Scholar] [CrossRef]
- Tang, M.S.L.; Shiu, S.C.C.; Godonoga, M.; Cheung, Y.W.; Liang, S.; Dirkzwager, R.M.; Kinghorn, A.B.; Fraser, L.A.; Heddle, J.G.; Tanner, J.A. An aptamer-enabled DNA nanobox for protein sensing. Nanomed. Nanotechnol. 2018, 14, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Godonoga, M.; Lin, T.Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.S.; Kuzuya, A.; et al. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, L.A.; Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.W.; Lim, B.; Shiu, S.C.; Tang, M.S.L.; Andrew, D.; Manitta, J.; et al. A portable microfluidic Aptamer-Tethered Enzyme Capture (APTEC) biosensor for malaria diagnosis. Biosens. Bioelectron. 2018, 100, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Dirkzwager, R.M.; Liang, S.L.; Tanner, J.A. Development of aptamer-based point-of-care diagnostic devices for malaria using three-dimensional printing rapid prototyping. ACS Sens. 2016, 1, 420–426. [Google Scholar] [CrossRef]
- Pei, H.; Zuo, X.; Zhu, D.; Huang, Q.; Fan, C. Functional DNA nanostructures for theranostic applications. Acc. Chem. Res. 2014, 47, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Wang, J.; Zhou, G.; Wang, J.; Wu, N.; Lu, J.; Gao, J.; Chen, X.; Shi, J.; Zuo, X.; et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. 2015, 54, 2151–2155. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Li, X.; Cui, F.; Qiu, Q.; Chen, X.; Huang, H. Aflatoxin B1 electrochemical aptasensor based on tetrahedral DNA nanostructures functionalized three dimensionally ordered macroporous MoS2–AuNPs film. ACS Appl. Mater. Interface 2018, 10, 17551–17559. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Song, P.; Zhou, G.B.; Zuo, X.L.; Aldalbahi, A.; Lou, X.D.; Shi, J.Y.; Fan, C.H. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 2016, 11, 1244–1263. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A Three-Dimensional Editing Tool for Complex DNA Structures; Springer: Berlin/Heidelberg, Germany, 2009; pp. 90–101. [Google Scholar]
- Wen, Y.; Pei, H.; Wan, Y.; Su, Y.; Huang, Q.; Song, S.; Fan, C. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal. Chem. 2011, 83, 7418–7423. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Wang, Z.; Meng, Z.; Wang, P.; San, L.; Wang, W.; Aldalbahi, A.; Li, L.; Shen, J.; Mi, X. DNA tetrahedral nanostructure-based electrochemical miRNA biosensor for simultaneous detection of multiple miRNAs in pancreatic carcinoma. ACS Appl. Mater. Interface 2017, 9, 24118–24125. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Wang, L.; Xu, L.; Li, L.; Ren, S.; Cao, C.; Jia, N.; Aldalbahi, A.; Song, S.; Shi, J.; et al. Electrochemical detection of PCR amplicons of Escherichia coli genome based on DNA nanostructural probes and polyHRP enzyme. Analyst 2016, 141, 5304–5310. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, R.; Zhu, J.; Lu, X.; Li, Y.; Qiu, S.; Jia, L.; Jiao, X.; Song, S.; Fan, C.; et al. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of Avian influenza A (H7N9) virus. ACS Appl. Mater. Interface 2015, 7, 8834–8842. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Lin, M.; Wang, P.; Pei, H.; Yan, J.; Shi, J.; Huang, Q.; He, D.; Fan, C.; Zuo, X. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem. 2014, 86, 2124–2130. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.X.; Huang, K.J.; He, L.L.; Wang, Y.H. Tetrahedral DNA probe coupling with hybridization chain reaction for competitive thrombin aptasensor. Biosens. Bioelectron. 2018, 100, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.M.; Zhou, Z.; Li, M.X.; Zhao, W.; Xu, J.J.; Chen, H.Y. DNA tetrahedral scaffolds-based platform for the construction of electrochemiluminescence biosensor. Biosens. Bioelectron. 2017, 90, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Wang, B.; Chen, X.; Li, X.; Tang, Y. Tetrahedral DNA nanostructure-based microRNA biosensor coupled with catalytic recycling of the analyte. ACS Appl. Mater. Interface 2015, 7, 6238–6243. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, B.; Wang, D.; Wen, Y.; Liu, G.; Dong, H.; Song, S.; Fan, C. DNA nanostructure-based universal microarray platform for high-efficiency multiplex bioanalysis in biofluids. ACS Appl. Mater. Interface 2014, 6, 17944–17953. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Giovanni, M.; Xie, J.P.; Fan, C.H.; Leong, D.T. Ultrasensitive IgG quantification using DNA nano-pyramids. NPG Asia Mater. 2014, 6, e112. [Google Scholar] [CrossRef]
- Abi, A.; Lin, M.H.; Pei, H.; Fan, C.H.; Ferapontova, E.E.; Zuo, X.L. Electrochemical switching with 3D DNA tetrahedral nanostructures self-assembled at gold electrodes. ACS Appl. Mater. Interface 2014, 6, 8928–8931. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.; Hao, P.; Guangbao, Y.; Lihua, W.; Shao, S.; Jie, C.; Lianhui, W.; Ali, A.; Shiping, S.; Jiye, S.; et al. A surface-confined proton-driven DNA pump using a dynamic 3D DNA scaffold. Adv. Mater. 2016, 28, 6860–6865. [Google Scholar]
- Zeng, D.; Zhang, H.; Zhu, D.; Li, J.; San, L.; Wang, Z.; Wang, C.; Wang, Y.; Wang, L.; Zuo, X.; et al. A novel ultrasensitive electrochemical DNA sensor based on double tetrahedral nanostructures. Biosens. Bioelectron. 2015, 71, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shuai, Z.H.; Zhou, H.; Luo, Z.M.; Liu, B.; Zhang, Y.N.; Zhang, L.; Chen, S.F.; Chao, J.; Weng, L.M.; et al. Single-molecule analysis of microRNA and logic operations using a smart plasmonic nanobiosensor. J. Am. Chem. Soc. 2018, 140, 3988–3993. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ge, Z.; Mao, X.; Zhou, G.; Zuo, X.; Shen, J.; Shi, J.; Li, J.; Wang, L.; Chen, X.; et al. Valency-controlled framework nucleic acid signal amplifiers. Angew. Chem. Int. Ed. 2018, 57, 7131–7135. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the oligonucleotides and protein are not available from the authors but it can be purchased from the manufacturers or purified as mentioned in the materials sections. |

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiu, S.C.-C.; Fraser, L.A.; Ding, Y.; Tanner, J.A. Aptamer Display on Diverse DNA Polyhedron Supports. Molecules 2018, 23, 1695. https://doi.org/10.3390/molecules23071695
Shiu SC-C, Fraser LA, Ding Y, Tanner JA. Aptamer Display on Diverse DNA Polyhedron Supports. Molecules. 2018; 23(7):1695. https://doi.org/10.3390/molecules23071695
Chicago/Turabian StyleShiu, Simon Chi-Chin, Lewis A. Fraser, Yifan Ding, and Julian A. Tanner. 2018. "Aptamer Display on Diverse DNA Polyhedron Supports" Molecules 23, no. 7: 1695. https://doi.org/10.3390/molecules23071695