Application of the Fluorescent Dye BODIPY in the Study of Lipid Dynamics of the Rice Blast Fungus Magnaporthe oryzae
Abstract
:1. Introduction
2. Results
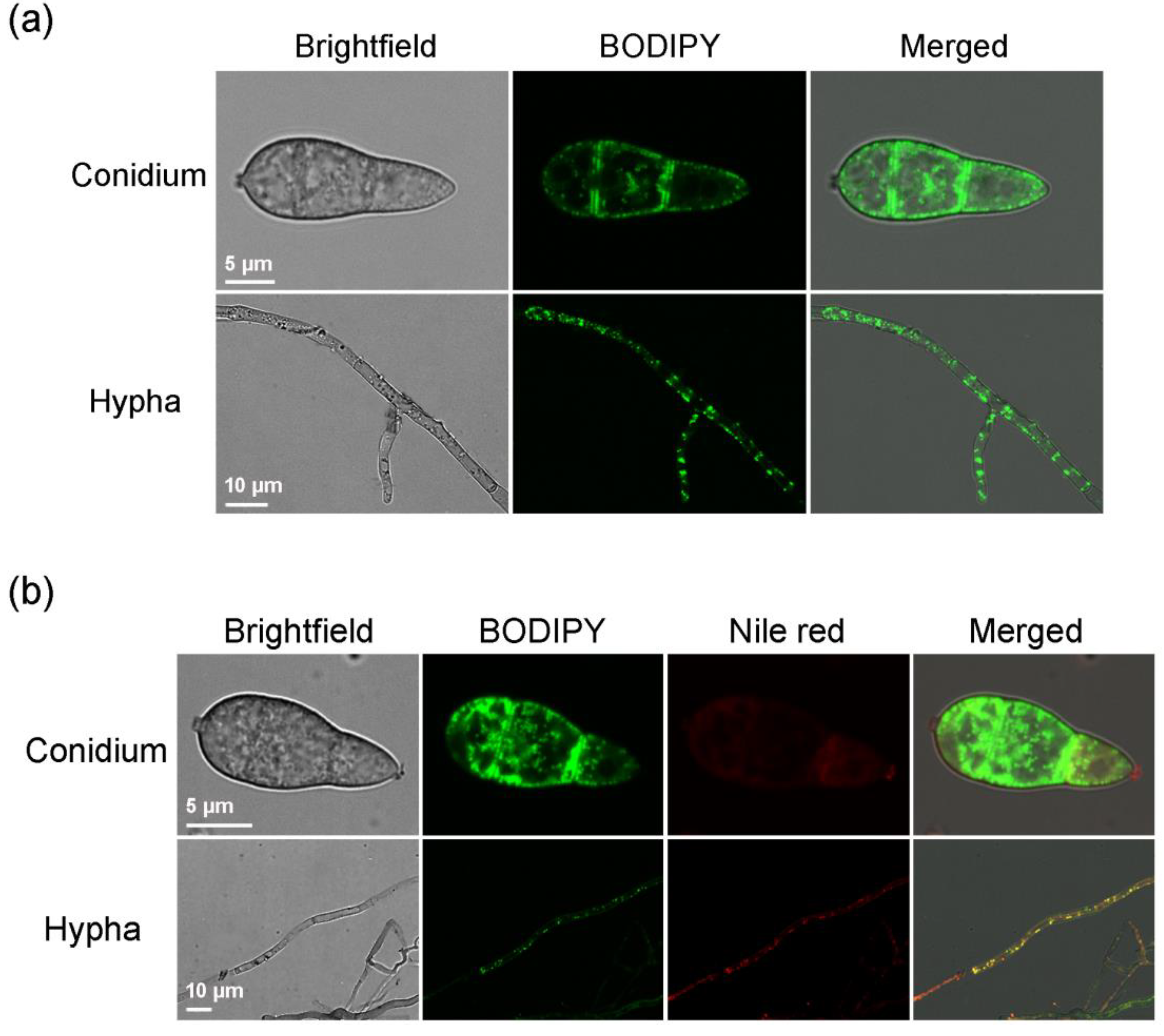
2.1. BODIPY Can Label Lipid Droplets in M. oryzae Mycelia and Conidia
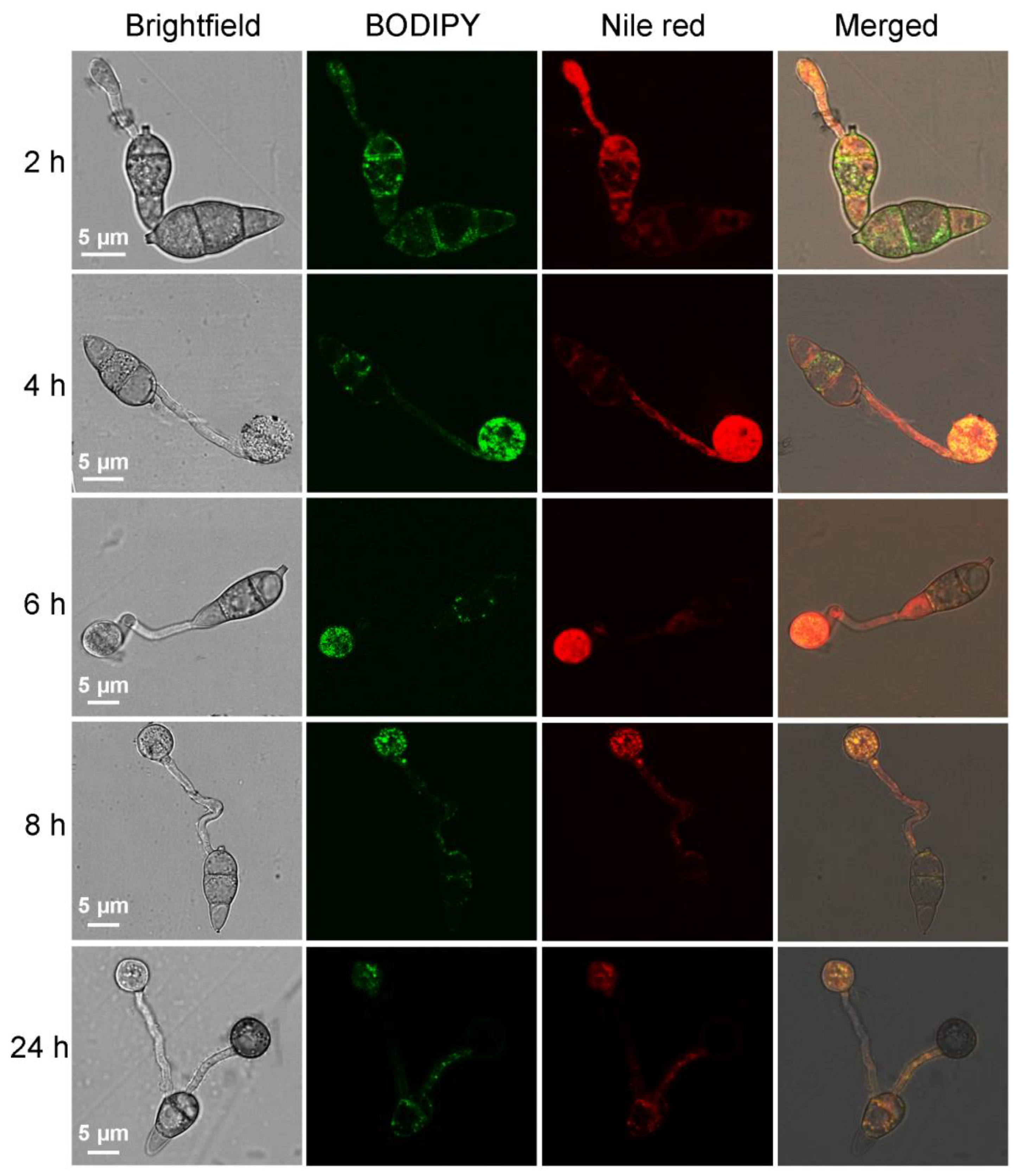
2.2. Using BODIPY to Trace the Lipid Migration during M. oryzae Conidium Germination and Appressorium Formation
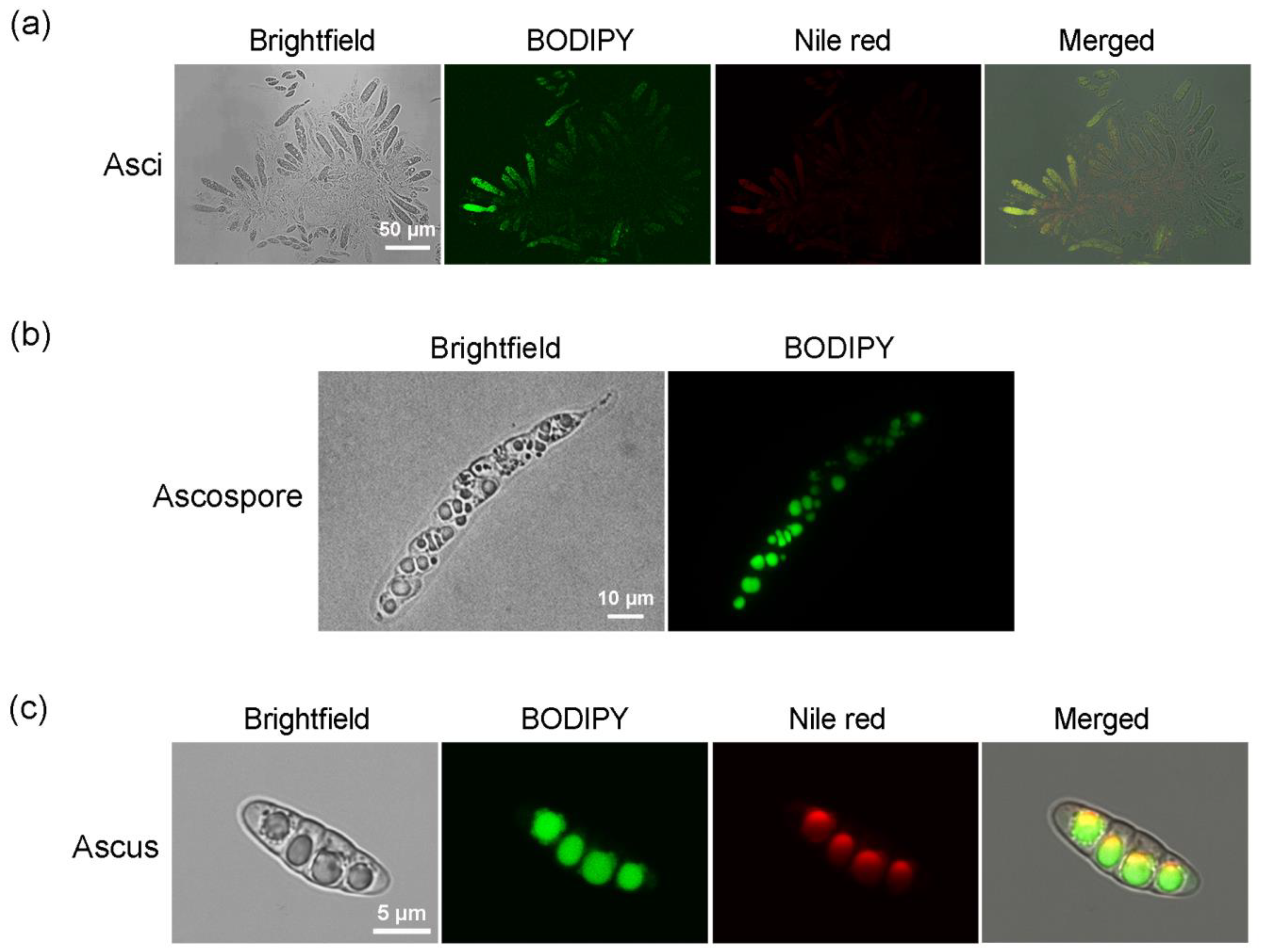
2.3. Labeling Lipids in Sexual Reproductive Structures of M. oryzae with BODIPY
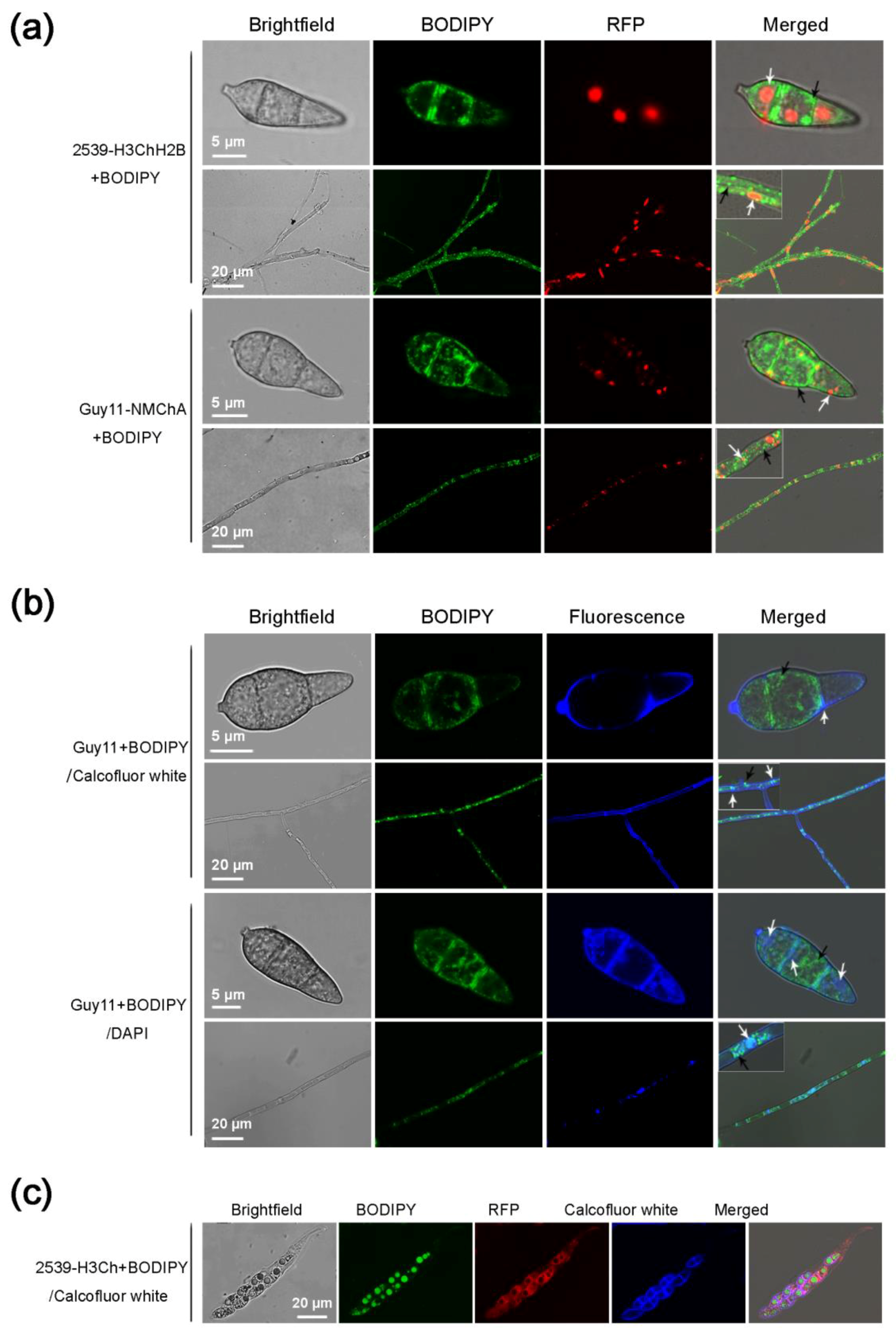
2.4. Using BODIPY in Combination with Fluorescent Proteins and Other Fluorescent Dyes in M. oryzae
3. Discussion
4. Materials and Methods
4.1. Strains, Media and Culture Conditions
4.2. Reagents and Preparation
4.3. Tissue Collection and Staining
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Skamnioti, P.; Gurr, S.J. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Hebert, T.T. The perfect stage of Pyricularia grisea. Phytopathology 1971, 61, 83–87. [Google Scholar] [CrossRef]
- Yoder, O.C.; Valent, B.; Chumley, F.G. Genetic nomenclature and practice for plant pathogenic fungi. Phytopathology 1986, 76, 383–385. [Google Scholar] [CrossRef]
- Talbot, N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J. Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 2007, 45, 437–456. [Google Scholar] [CrossRef] [PubMed]
- Valent, B.; Chumley, F.G. Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu. Rev. Phytopathol. 1991, 29, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Dejong, J.C.; Mccormack, B.J.; Smirnoff, N.; Talbot, N.J. Glycerol generates turgor in rice blast. Nature 1997, 389, 244–245. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Soanes, D.M.; Kershaw, M.J.; Talbot, N.J. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol. Plant-Microbe Interact. 2007, 20, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Bhambra, G.K.; Wang, Z.Y.; Soanes, D.M.; Wakley, G.E.; Talbot, N.J. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 2006, 61, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Thines, E.; Weber, R.W.; Talbot, N.J. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 2000, 12, 1703–1718. [Google Scholar] [PubMed]
- Wang, J.; Zhang, Z.; Wang, Y.; Li, L.; Chai, R.; Mao, X.; Jiang, H.; Qiu, H.; Du, X.; Lin, F.; et al. PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae. PLoS ONE 2013, 8, e55554. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, J.; Zhang, Z.; Wang, Y.; Liu, M.; Jiang, H.; Chai, R.; Mao, X.; Qiu, H.; Liu, F.; et al. MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS ONE 2014, 9, e85252. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Gu, Z.; Yang, N.; Li, L.; Yue, X.; Que, Y.; Sun, G.; Wang, Z.; Wang, J. Identification and characterization of the peroxin 1 gene MoPEX1 required for infection-related morphogenesis and pathogenicity in Magnaporthe oryzae. Sci. Rep. 2016, 6, 36292. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, P.; Fowler, S.D. Spectrofluorometric studies of the lipid probe, Nile red. J. Lipid Res. 1985, 26, 781–789. [Google Scholar] [PubMed]
- Fowler, S.D.; Brown, W.J.; Warfel, J.; Greenspan, P. Use of Nile red for the rapid in situ quantitation of lipids on thin-layer chromatograms. J. Lipid Res. 1987, 28, 1225–1232. [Google Scholar] [PubMed]
- Priscu, J.C.; Priscu, L.R.; Palmisano, A.C.; Sullivan, C.W. Estimation of neutral lipid levels in antarctic sea ice microalgae by nile red fluorescence. Antarct. Sci. 1990, 2, 149–155. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. Lipoprotein metabolism in the macrophage: Implications for cholesterol deposition in atherosclerosis. Annu. Rev. Biochem. 1983, 52, 223–261. [Google Scholar] [CrossRef] [PubMed]
- Siegler, H.D.L.H.; Ayidzoe, W.; Ben-Zvi, A.; Burrell, R.E.; Mccaffrey, W.C. Improving the reliability of fluorescence-based neutral lipid content measurements in microalgal cultures. Algal Res. 2012, 1, 176–184. [Google Scholar] [CrossRef]
- Kobae, Y.; Gutjahr, C.; Paszkowski, U.; Kojima, T.; Fujiwara, T.; Hata, S. Lipid droplets of arbuscular mycorrhizal fungi emerge in concert with arbuscule collapse. Plant Cell Physiol. 2014, 55, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Yamaoka, M.; Kamisaka, Y. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. J. Microbiol. Methods 2004, 56, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, I.; Krishnamoorthy, G. Probing the link between proton transport and water content in lipid membranes. J. Phys. Chem. B 2001, 105, 1484–1488. [Google Scholar]
- Karolin, J.; Johansson, L.; Strandberg, L.; Ny, T. Fluorescence and absorption spectroscopic properties of Dipyrrometheneboron Difluoride (BODIPY) derivatives in liquids, lipid membranes, and proteins. J. Am. Chem. Soc. 1994, 116, 7801–7806. [Google Scholar] [CrossRef]
- Laurens, L.M.L.; Wolfrum, E.J. Feasibility of Spectroscopic Characterization of algal lipids: Chemometric correlation of NIR and FTIR spectra with exogenous lipids in algal biomass. Bioenergy Res. 2011, 4, 22–35. [Google Scholar] [CrossRef]
- Elle, I.C.; Olsen, L.C.B.; Pultz, D.; Rødkær, S.V.; Færgeman, N.J. Something worth dyeing for: Molecular tools for the dissection of lipid metabolism in Caenorhabditis elegans. FEBS Lett. 2010, 584, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Li, P.; Li, P.; Xie, T.; Yu, F.; Tang, B. The synthesis of polarity-sensitive fluorescent dyes based on the BODIPY chromophore. Dyes Pigments 2011, 89, 217–222. [Google Scholar] [CrossRef]
- Niu, S.L.U.; Ulrich, G.; Retailleau, P.; Harrowfield, J.; Ziessel, R. New insights into the solubilization of Bodipy dyes. Tetrahedron Lett. 2009, 50, 3840–3844. [Google Scholar] [CrossRef]
- Yang, L.; Simionescu, R.; Lough, A.; Yan, H. Some observations relating to the stability of the BODIPY fluorophore under acidic and basic conditions. Dyes Pigments 2011, 91, 264–267. [Google Scholar] [CrossRef]
- Loudet, A.; Burgess, K. BODIPY® Dyes and Their Derivatives: Syntheses and Spectroscopic Properties. In Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine; World Scientific Publishing: Singapore, 2010; Volume 8, pp. 1–164. [Google Scholar]
- Gocze, P.M.; Freeman, D.A. Factors underlying the variability of Lipid droplet fluorescence in MA-10 leydig tumor cells. Cytometry 1994, 17, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.S.; Hardin, W.R.; Petersen, T.W.; Cattolico, R.A. Visualizing “green oil” in live algal cells. J. Biosci. Bioeng. 2010, 109, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, N.; Sung, M.; Yim, S.S.; Park, M.S.; Yang, J.W.; Jeong, K.J. Evaluation of intracellular lipid bodies in Chlamydomonas reinhardtii strains by flow cytometry. Bioresour. Technol. 2013, 138, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhou, J.; He, Y.; Xie, Q.; Chen, A.; Zheng, H.; Shi, L.; Zhao, X.; Zhang, C.; Huang, Q.; et al. Retromer is essential for autophagy-dependent plant infection by the rice blast fungus. PLoS Genet. 2015, 11, e1005704. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Hu, J.; Meng, X.; Zhang, Q.; Zhu, K.; Chen, X.; Chen, X.; Li, G.; Wang, Z.; et al. Functional characterization of electron-transferring flavoprotein and its dehydrogenase required for fungal development and plant infection by the rice blast fungus. Sci. Rep. 2016, 6, 24911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Thornton, C.R.; Kershaw, M.J.; Debao, L.; Talbot, N.J. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 2003, 47, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu. Rev. Biochem. 1977, 46, 897–930. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, S.R. The comparative histologic application of phorwite Fabric brighteners. J. Histotechnol. 1989, 12, 43–45. [Google Scholar] [CrossRef]
- Monheit, J.E.; Cowan, D.F.; Moore, D.G. Rapid detection of fungi in tissues using calcofluor white and fluorescence microscopy. Arch. Pathol. Lab. Med. 1984, 108, 616–618. [Google Scholar] [PubMed]
- Kubista, M.; Akerman, B.; Nordén, B. Characterization of interaction between DNA and 4′,6-diamidino-2-phenylindole by optical spectroscopy. Biochemistry 1987, 26, 4545. [Google Scholar] [CrossRef] [PubMed]
- Odenbach, D.; Breth, B.; Thines, E.; Weber, R.W.; Anke, H.; Foster, A. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 2007, 64, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Tanious, F.A.; Veal, J.M.; Buczak, H.; Ratmeyer, L.S.; Wilson, W.D. DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: Minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 1992, 31, 3103–3112. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Li, L.; Lao, L.; Liu, Y.; Wei, Q.; Ji, Q.; Sun, G.; Lin, F.; Wang, J.; Bao, G. Application of the red fluorescent protein mCherry in mycelial labeling and organelle tracing in the dermatophyte Trichophyton mentagrophytes. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Talbot, N.J.; Ebbole, D.J.; Hamer, J.E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 1993, 5, 1575–1590. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Xue, C.; Zheng, L.; Lam, S.; Xu, J.R. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 2002, 15, 183–192. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are commercially available from the companies referred. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Guo, X.; Li, L.; Qiu, H.; Zhang, Z.; Wang, Y.; Sun, G. Application of the Fluorescent Dye BODIPY in the Study of Lipid Dynamics of the Rice Blast Fungus Magnaporthe oryzae. Molecules 2018, 23, 1594. https://doi.org/10.3390/molecules23071594
Wang J, Guo X, Li L, Qiu H, Zhang Z, Wang Y, Sun G. Application of the Fluorescent Dye BODIPY in the Study of Lipid Dynamics of the Rice Blast Fungus Magnaporthe oryzae. Molecules. 2018; 23(7):1594. https://doi.org/10.3390/molecules23071594
Chicago/Turabian StyleWang, Jiaoyu, Xiaoyu Guo, Ling Li, Haiping Qiu, Zhen Zhang, Yanli Wang, and Guochang Sun. 2018. "Application of the Fluorescent Dye BODIPY in the Study of Lipid Dynamics of the Rice Blast Fungus Magnaporthe oryzae" Molecules 23, no. 7: 1594. https://doi.org/10.3390/molecules23071594



