Sterepinic Acids A–C, New Carboxylic Acids Produced by a Marine Alga-Derived Fungus
Abstract
:1. Introduction
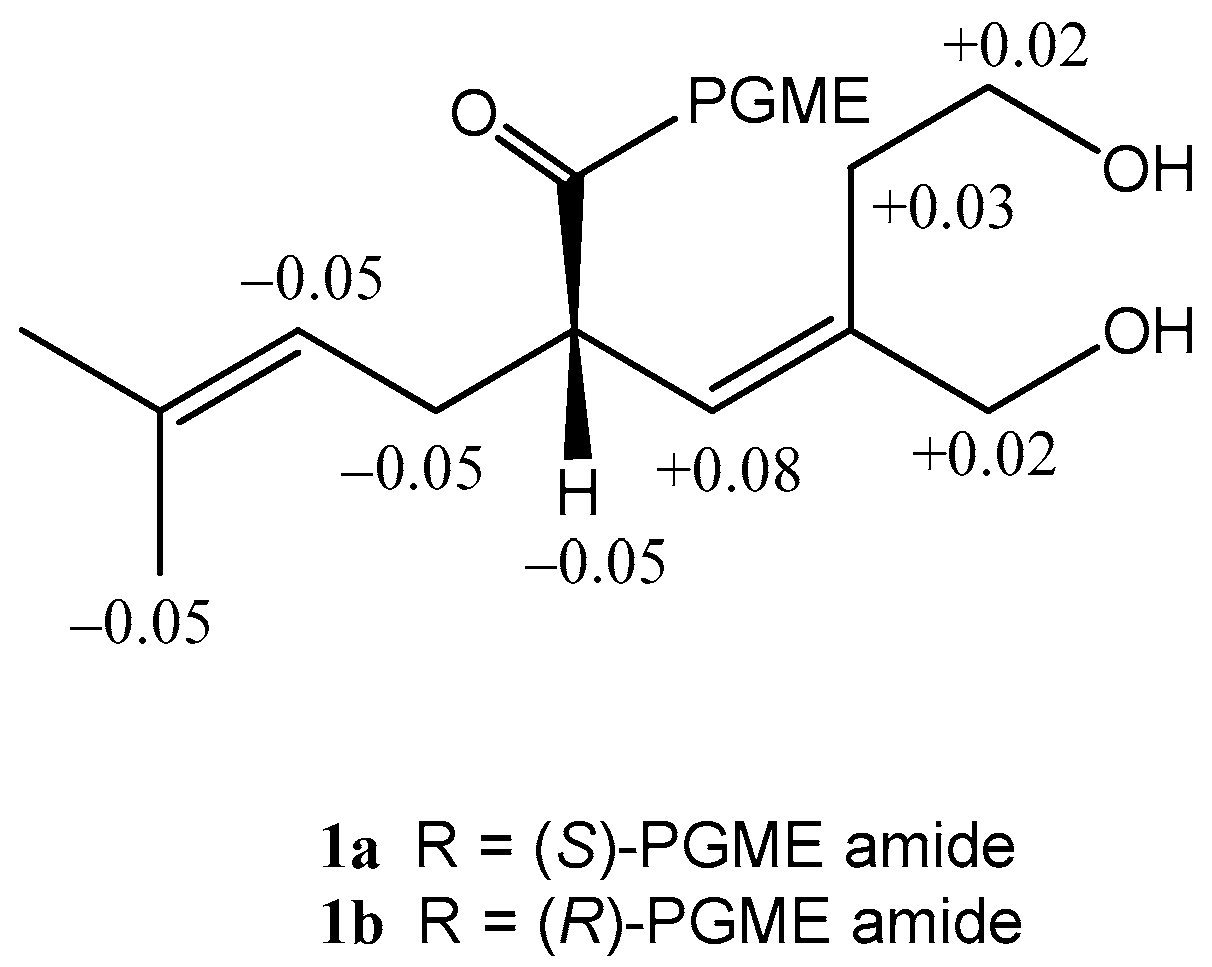
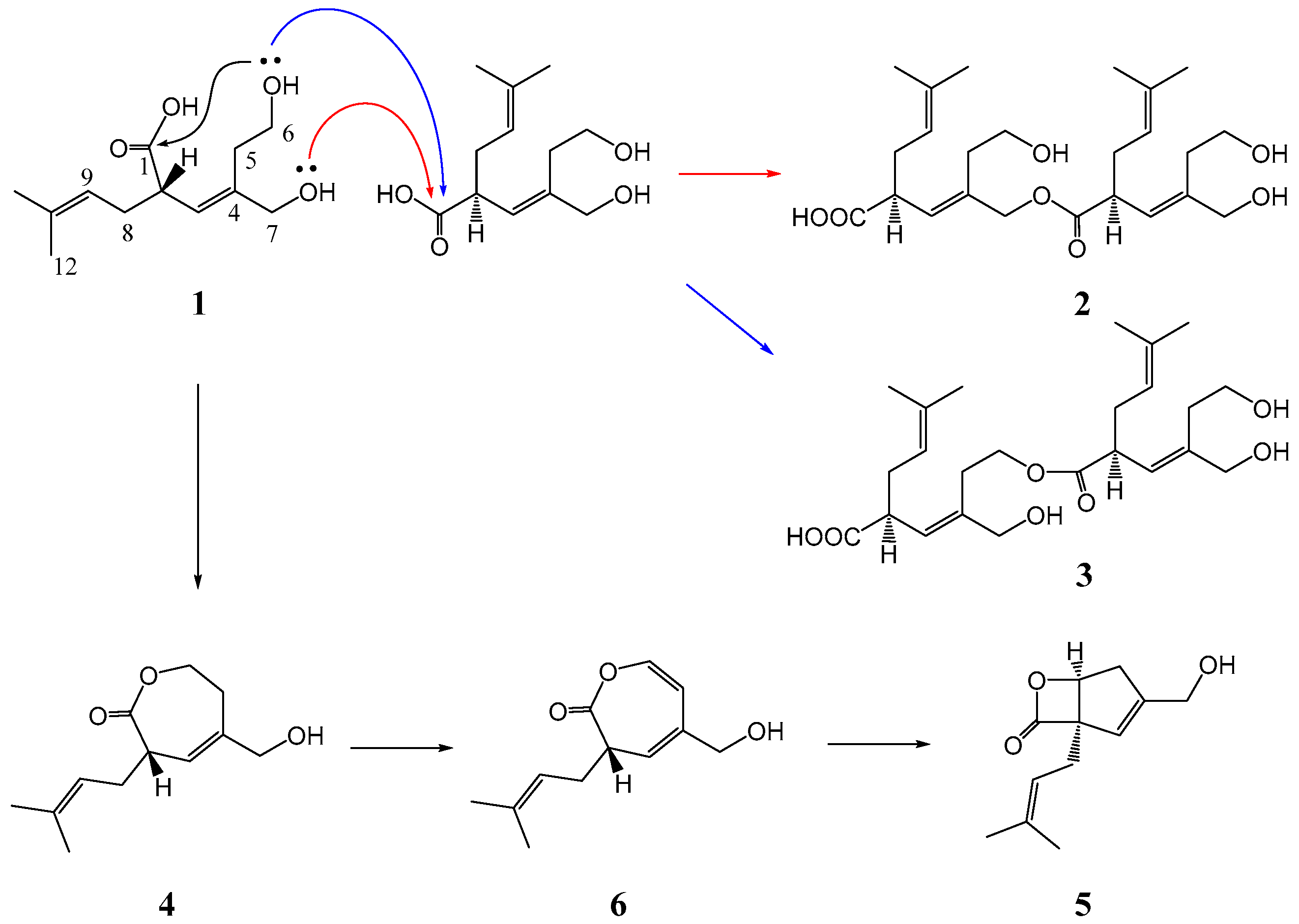
2. Results
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Culturing and Isolation of Metabolites
3.4. Chemical Transformation
3.4.1. Formation of the (S)- and (R)-PGME Amides
3.4.2. Formation of Methyl Ester of 1
3.4.3. Methanolysis of 2–4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muroga, Y.; Yamada, T.; Numata, A.; Tanaka, R. Chaetomugilins I–O, new potent cytotoxic metabolites from a marine-fish-derived Chaetomium species. Stereochemistry and biological activities. Tetrahedron 2009, 65, 7580–7586. [Google Scholar] [CrossRef]
- Yamada, T.; Kitada, H.; Kajimoto, T.; Numata, A.; Tanaka, R. The relationship between the CD Cotton effect and the absolute configuration of FD-838 and its seven stereoisomers. J. Org. Chem. 2010, 75, 4146–4153. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Kikuchi, T.; Tanaka, R.; Numata, A. Halichoblelides B and C, potent cytotoxic macrolides from a Streptomyces species separated from a marine fish. Tetrahedron Lett. 2012, 53, 2842–2846. [Google Scholar] [CrossRef]
- Kitano, M.; Yamada, T.; Amagata, T.; Minoura, K.; Tanaka, R.; Numata, A. Novel pyridinopyrone sesquiterpene type pileotin produced by a sea urchin-derived Aspergillus sp. Tetrahedron Lett. 2012, 53, 4192–4194. [Google Scholar] [CrossRef]
- Yamada, T.; Mizutani, Y.; Umebayashi, Y.; Inno, N.; Kawashima, M.; Kikuchi, T.; Tanaka, R. A novel ketoaldehyde decalin derivative, produced by a marine sponge-derived Trichoderma harzianum. Tetrahedron Lett. 2014, 55, 662–664. [Google Scholar] [CrossRef]
- Yamada, T.; Umebayashi, Y.; Kawashima, M.; Sugiura, Y.; Kikuchi, T.; Tanaka, R. Determination of the chemical structures of tandyukisins B–D, isolated from a marine sponge-derived fungus. Mar. Drugs 2015, 13, 3231–3240. [Google Scholar] [CrossRef] [PubMed]
- Suzue, M.; Kikuchi, T.; Tanaka, R.; Yamada, T. Tandyukisins E and F, novel cytotoxic decalin derivatives isolated from a marine sponge-derived fungus. Tetrahedron Lett. 2016, 57, 5070–5073. [Google Scholar] [CrossRef]
- Yamada, T.; Suzue, M.; Arai, T.; Kikuchi, T.; Tanaka, R. Trichodermanins C–E, new diterpenes with a fused 6-5-6-6 ring system produced by a marine sponge-derived fungus. Marine Drugs 2017, 15, 169. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Z.; Wang, F.; Liao, T.G.; Tang, J.G.; Steglich, W.; Zhu, H.J.; Liu, J.K. Vibralactone: A lipase inhibitor with an unusual fused β-lactone produced by cultures of the basidiomycete. Boreostereum vibrans. Org. Lett. 2006, 8, 5749–5752. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.Y.; Wang, F.; Yang, X.L.; Fang, L.Z.; Dong, Z.J.; Zhu, H.J.; Liu, J.K. Derivatives of vibralactone from cultures of the basidiomycete Boreostereum vibrans. Chem. Pharm. Bull. 2008, 56, 1286–1288. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.Y.; Zhang, L.; Dong, Z.J.; Yang, Z.L.; Leng, Y.; Liu, J.K. Vibralactones D–F from cultures of the basidiomycete Boreostereum vibrans. Chem. Pharm. Bull. 2010, 58, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.H.; Feng, T.; Li, Z.H.; Li, L.; Liu, J.K. Twelve new compounds from the basidiomycete Boreostereum vibrans. Nat. Prod. Bioprospect. 2012, 2, 200–205. [Google Scholar] [CrossRef]
- Wang, G.Q.; Wei, K.; Feng, T.; Li, Z.H.; Zhang, L.; Wang, Q.A.; Liu, J.K. Vibralactones G-J from cultures of the basidiomycete Boreostereum vibrans. J. Asian Nat. Prod. Res. 2012, 14, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Wei, K.; Li, Z.H.; Feng, T.; Ding, J.H.; Wang, Q.A.; Liu, J.K. Three new compounds from the cultures of basidiomycete Boreostereum vibrans. J. Asian Nat. Prod. Res. 2013, 15, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Zhao, Z.Z.; Yin, R.H.; Yin, X.; Feng, T.; Li, Z.H.; Wei, K.; Liu, J.K. Six new vibralactone derivatives from cultures of the fungus Boreostereum vibrans. Nat. Prod. Bioprospect. 2014, 4, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, T.; Kusumi, T. Phenylglycine Methyl Ester, a Useful Tool for Absolute Configuration Determination of various chiral carboxylic acids. J. Org. Chem. 2000, 65, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.J.; Yang, Y.L.; Du, L.; Liu, J.K.; Zeng, Y. Elucidating the biosynthetic pathway for vibralactone: A pancreatic lipase inhibitor with a fused bicyclic β-lactone. Angew. Chem. Int. Ed. 2013, 52, 2298–2302. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
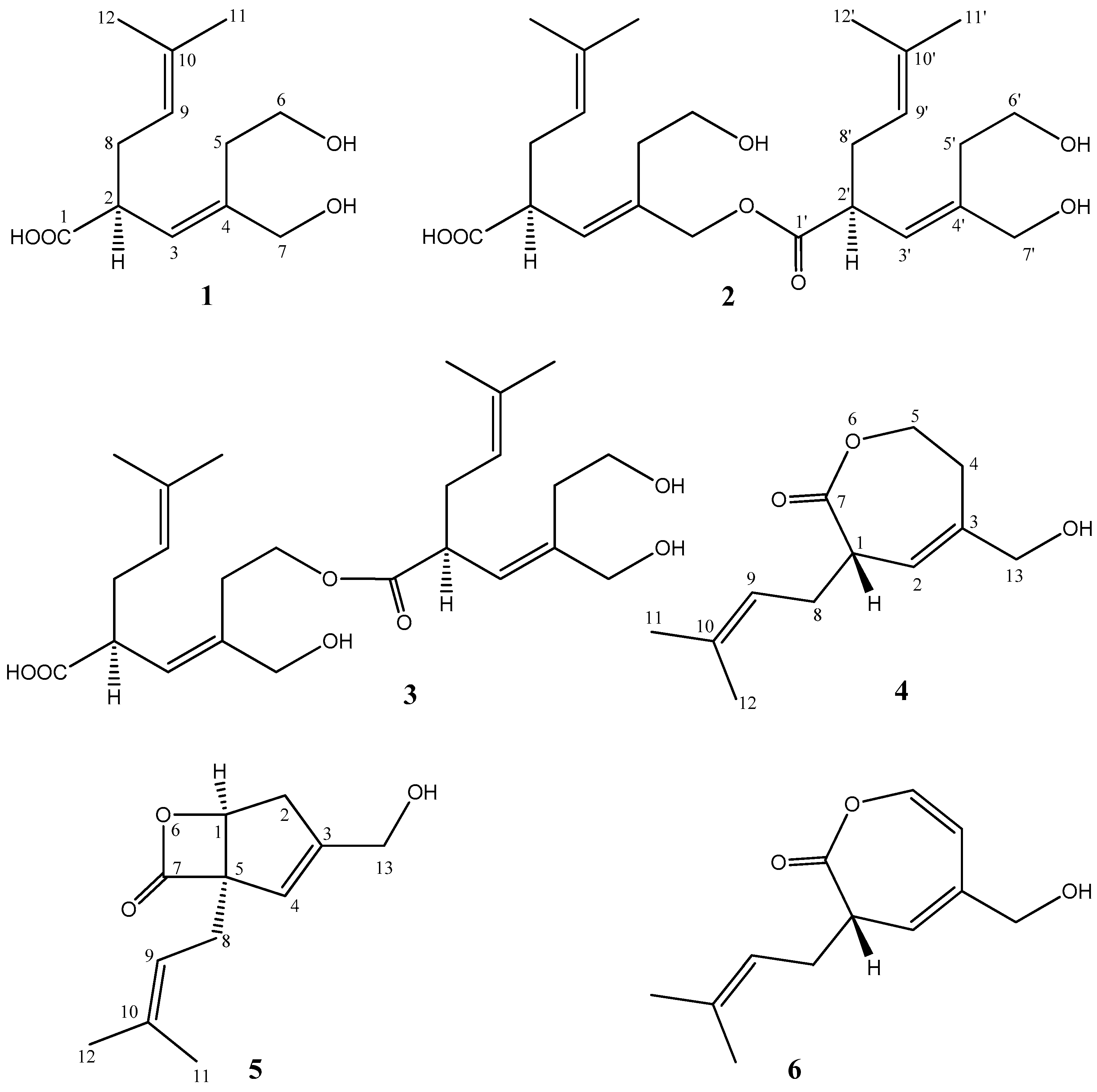
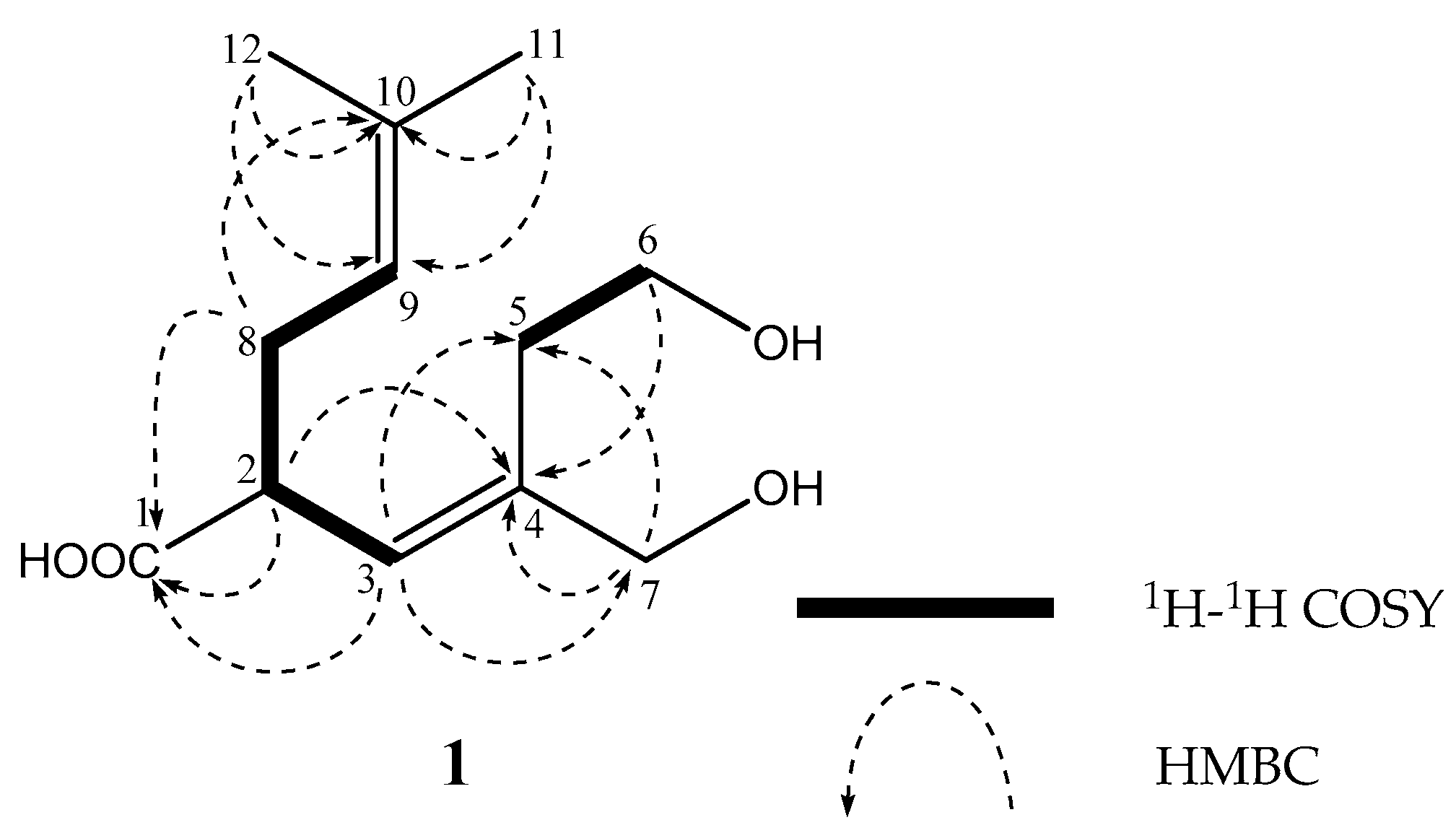
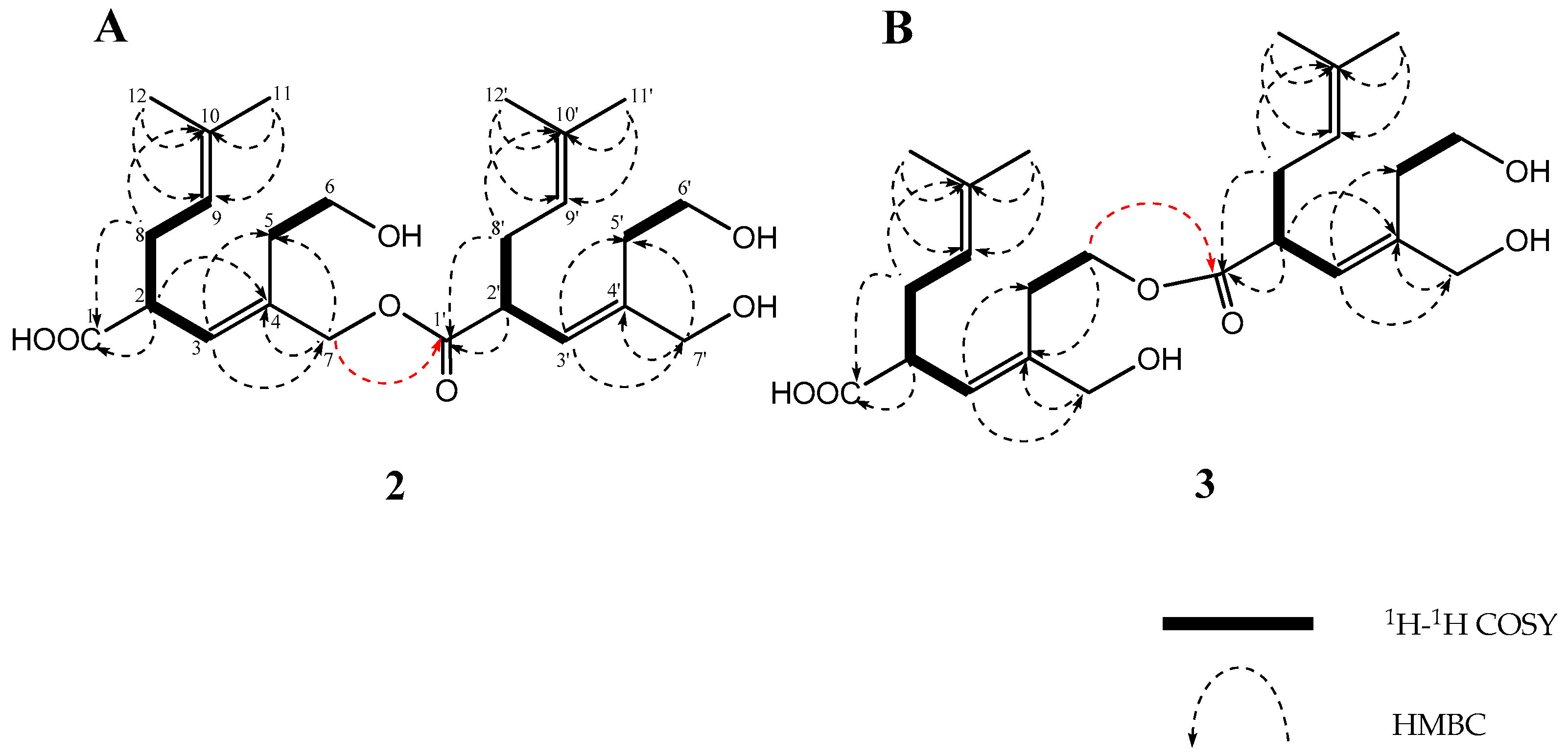
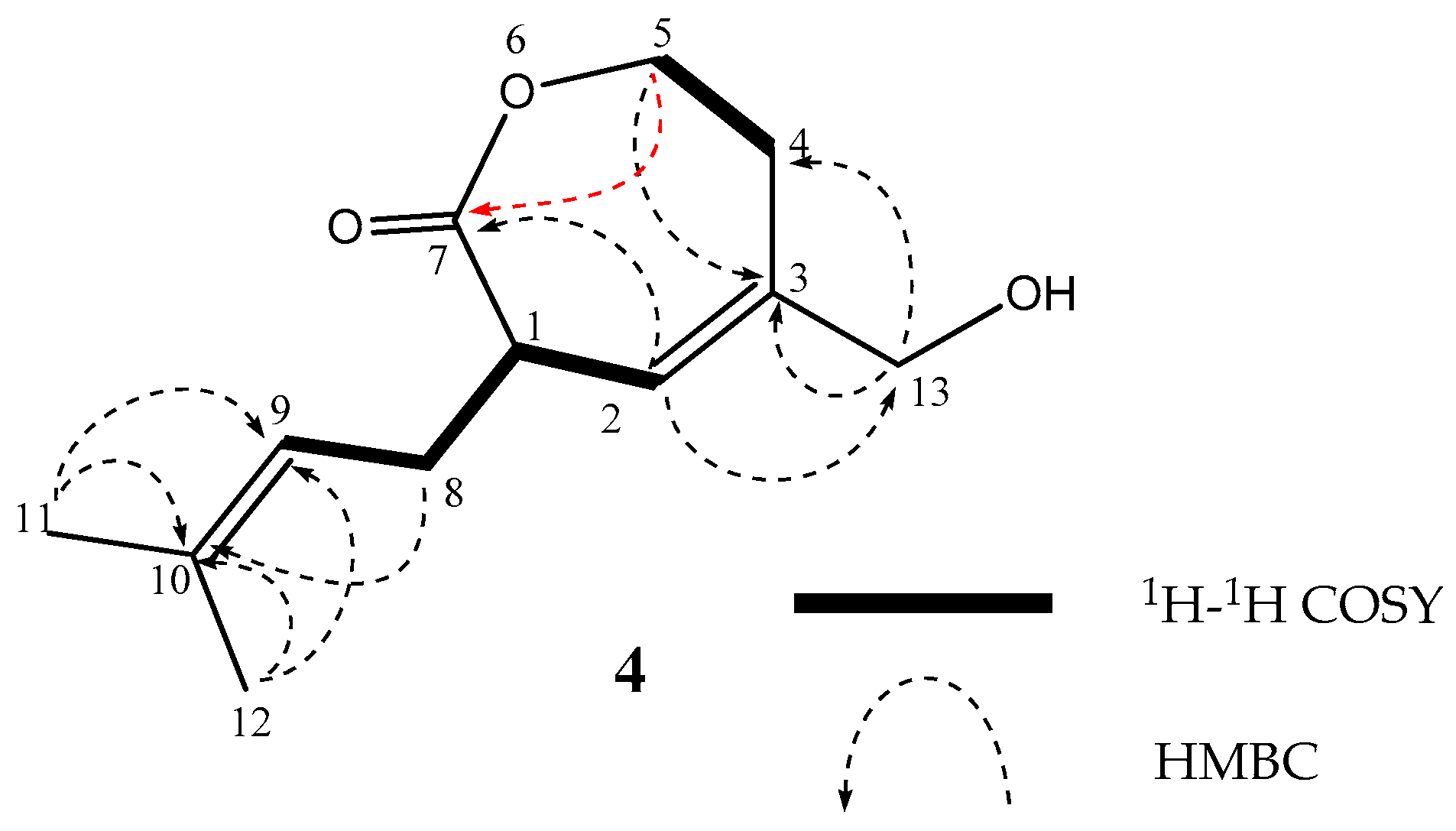
| Position | 1 | 2 | 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δHa | δC | δHa | δC | δHa | δC | ||||||||
| 1 | 177.5 | (s) | 173.5 | (s) | 174.3 | (s) | |||||||
| 2 | 3.27 | m | 44.9 | (d) | 3.28 | m | 45.4 | (d) | 3.28 | m | 44.9 | (d) | |
| 3 | 5.50 | d (10.2) | 127.0 | (d) | 5.49 | d (10.8) | 129.3 | (d) | 5.55 | d (9.6) | 129.3 | (d) | |
| 4 | 138.7 | (s) | 133.9 | (s) | 137.9 | (s) | |||||||
| 5A | 2.25 | m | 31.9 | (t) | 2.18 | m | 32.3 | (t) | 2.30 | ddd (14.4, 5.4, 5.4) | 27.7 | (t) | |
| 5B | 2.49 | m | 2.54 | m | 2.54 | ddd (14.4, 5.4, 5.4) | |||||||
| 6A | 3.68 | br s | 61.0 | (t) | 3.65 | br s | 61.4 | (t) | 4.20 | m | 63.5 | (t) | |
| 6B | 66.8 | (t) | 3.72 | br s | |||||||||
| 7A | 4.03 | br s | 4.05 | d (13.2) | 67.9 | (t) | 4.07 | m | 66.5 | (t) | |||
| 7B | d (13.2) | ||||||||||||
| 8A | 2.20 | m | 30.9 | (t) | 2.20 | m | 30.8 b5 | (t) | 2.20 | m | 31.4 | (t) | |
| 8B | 2.44 | m | 2.46 | m | 2.44 | m | |||||||
| 9 | 5.04 | dd | 120.2 | (d) | 5.03 | m | 120.2 b6 | (d) | 5.02 b1 | dd (7.2, 7.2) | 120.2 b2 | (d) | |
| 10 | 134.1 | (s) | 134.2 b7 | (s) | 134.3 | (s) | |||||||
| 11 | 1.67 | s | 25.7 | (q) | 1.67 | s | 25.7 | (q) | 1.67 | s | 25.7 | (q) | |
| 12 | 1.60 | s | 17.8 | (q) | 1.60 | s | 17.8 | (q) | 1.59 b3 | s | 17.8 b4 | (q) | |
| 1′ | 173.5 | (s) | 174.3 | (s) | |||||||||
| 2′ | 3.28 | m | 45.4 | (d) | 3.28 | m | 44.9 | (d) | |||||
| 3′ | 5.52 | d (10.8) | 127.2 | (d) | 5.51 | d (9.6) | 127.2 | (d) | |||||
| 4′ | 139.6 | (s) | 138.7 | (s) | |||||||||
| 5′A | 2.29 | m | 32.3 | (t) | 2.25 | m | 32.2 | (t) | |||||
| 5′B | 2.54 | m | 2.51 | m | |||||||||
| 6′A | 3.72 | br s | 60.5 | (t) | 3.65 | br s | 61.1 | (t) | |||||
| 6′B | 3.71 | br s | |||||||||||
| 7′ | 4.05 | br s | 67.5 | (t) | 4.02 | m | 67.4 | (t) | |||||
| 8′A | 2.20 | m | 30.6 b5 | (t) | 2.20 | m | 31.4 | (t) | |||||
| 8′B | 2.46 | m | 2.44 | m | |||||||||
| 9′ | 5.03 | m | 120.3 b6 | (d) | 5.06 b1 | dd (7.2, 7.2) | 120.3 b2 | (d) | |||||
| 10′ | 134.3 b7 | (s) | 134.3 | (s) | |||||||||
| 11′ | 1.67 | s | 25.7 | (q) | 1.67 | s | 25.7 | (q) | |||||
| 12′ | 1.60 | s | 17.8 | (q) | 1.61 b3 | s | 17.9 b4 | (q) | |||||
| Position | 4 | |||
|---|---|---|---|---|
| δHa | δC | |||
| 1 | 3.68 | m | 40.2 | (d) |
| 2 | 5.36 | br s | 121.2 | (d) |
| 3 | 139.2 | (s) | ||
| 4A | 2.45 | br d (19.2) | 30.3 | (t) |
| 4B | 2.59 | m | ||
| 5α | 4.68 | ddd (12.6, 12.6, 1.8) | 64.4 | (t) |
| 5β | 4.33 | ddd (12.6, 4.8, 2.4) | ||
| 6 | ||||
| 7 | 174.3 | (s) | ||
| 8A | 2.33 | ddd (14.4, 6.6, 6.6) | 30.1 | (t) |
| 8B | 2.52 | ddd (14.4, 6.6, 6.6) | ||
| 9 | 5.14 | dd (6.6, 6.6) | 120.9 | (d) |
| 10 | 134.6 | (s) | ||
| 11 | 1.72 | s | 25.8 | (q) |
| 12 | 1.67 | s | 18.0 | (q) |
| 13A | 3.99 | d (13.8) | 67.4 | (t) |
| 13B | 4.01 | d (13.8) | ||
| Compounds | Cell Line P388 | Cell Line HL-60 | Cell Line L1210 |
|---|---|---|---|
| IC50 (μM) a | IC50 (μM) a | IC50 (μM) a | |
| 1 | >500 | >500 | >500 |
| 2 | >500 | 236.7 | >500 |
| 3 | >500 | 60.2 | 480.9 |
| 4 | >500 | 189.5 | >500 |
| 5-fluorouracil b | 6 | 4.9 | 4.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, T.; Matsuda, M.; Seki, M.; Hirose, M.; Kikuchi, T. Sterepinic Acids A–C, New Carboxylic Acids Produced by a Marine Alga-Derived Fungus. Molecules 2018, 23, 1336. https://doi.org/10.3390/molecules23061336
Yamada T, Matsuda M, Seki M, Hirose M, Kikuchi T. Sterepinic Acids A–C, New Carboxylic Acids Produced by a Marine Alga-Derived Fungus. Molecules. 2018; 23(6):1336. https://doi.org/10.3390/molecules23061336
Chicago/Turabian StyleYamada, Takeshi, Miwa Matsuda, Mayuko Seki, Megumi Hirose, and Takashi Kikuchi. 2018. "Sterepinic Acids A–C, New Carboxylic Acids Produced by a Marine Alga-Derived Fungus" Molecules 23, no. 6: 1336. https://doi.org/10.3390/molecules23061336






