To Decipher the Mycoplasma hominis Proteins Targeting into the Endoplasmic Reticulum and Their Implications in Prostate Cancer Etiology Using Next-Generation Sequencing Data
Abstract
:1. Introduction
2. Results
2.1. Selection of the Whole Proteome Database of M. hominis
2.2. Prediction of the Subcellular Localization by cNLS Mapper
2.3. Prediction of ER Localization in M. hominis Proteins Using Hum-Mploc 3.0
3. Discussion
4. Materials and Methods
4.1. Proteins and Prediction Analysis
4.2. Choice of Protein Database for M. hominis
4.3. Prediction of the Subcellular Localization by cNLS Mapper
4.4. Prediction of ER Localization in M. hominis Proteins Using Hum-Mploc 3.0
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Eaton, K.; Yang, W. Registered report: Intestinal inflammation targets cancer-inducing activity of the microbiota. eLife 2015, 4, e04186. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Muhlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Barykova, Y.A.; Logunov, D.Y.; Shmarov, M.M.; Vinarov, A.Z.; Fiev, D.N.; Vinarova, N.A.; Rakovskaya, I.V.; Baker, P.S.; Shyshynova, I.; Stephenson, A.J.; et al. Association of Mycoplasma hominis infection with prostate cancer. Oncotarget 2011, 2, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Cavarretta, I.; Ferrarese, R.; Cazzaniga, W.; Saita, D.; Luciano, R.; Ceresola, E.R.; Locatelli, I.; Visconti, L.; Lavorgna, G.; Briganti, A.; et al. The Microbiome of the Prostate Tumor Microenvironment. Eur. Urol. 2017, 72, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, J.Y.; Wu, J.; Meng, L.; Shou, C.C. Mycoplasma infections and different human carcinomas. World J. Gastroenterol. 2001, 7, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar] [PubMed]
- Cimolai, N. Do mycoplasmas cause human cancer? Can. J. Microbiol. 2001, 47, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.H.; Tsai, S.; Rodriguez, J.; Lo, S.C. Mycoplasmal infections prevent apoptosis and induce malignant transformation of interleukin-3-dependent 32D hematopoietic cells. Mol. Cell. Biol. 1999, 19, 7995–8002. [Google Scholar] [CrossRef] [PubMed]
- Barykova Iu, A.; Shmarov, M.M.; Logunov, D.; Verkhovskaia, L.V.; Aliaev Iu, G.; Fiev, D.N.; Vinarov, A.Z.; Vinarova, N.A.; Rakovskaia, I.V.; Naroditskii, B.S.; et al. Identification of Mycoplasma in patients with suspected prostate cancer. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 2010, 4, 81–85. [Google Scholar]
- Khan, S.; Zakariah, M.; Rolfo, C.; Robrecht, L.; Palaniappan, S. Prediction of Mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget 2017, 8, 30830–30843. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.C.; Tsai, S. Mycoplasmas and human prostate cancer: An exciting but cautionary note. Oncotarget 2011, 2, 352–355. [Google Scholar] [PubMed]
- Caini, S.; Gandini, S.; Dudas, M.; Bremer, V.; Severi, E.; Gherasim, A. Sexually transmitted infections and prostate cancer risk: A systematic review and meta-analysis. Cancer Epidemiol. 2014, 38, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, A.; Nadel, B.; Raoult, D.; Neefjes, J.; Gorvel, J.P. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat. Rev. Microbiol. 2017, 15, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Imran, A.; Khan, A.A.; Abul Kalam, M.; Alshamsan, A. Systems Biology Approaches for the Prediction of Possible Role of Chlamydia pneumoniae Proteins in the Etiology of Lung Cancer. PLoS ONE 2016, 11, e0148530. [Google Scholar] [CrossRef] [PubMed]
- Khan, S. Potential role of Escherichia coli DNA mismatch repair proteins in colon cancer. Crit. Rev. Oncol. Hematol. 2015, 96, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Wear, D.J.; Shih, J.W.; Lo, S.C. Mycoplasmas and oncogenesis: Persistent infection and multistage malignant transformation. Proc. Natl. Acad. Sci. USA 1995, 92, 10197–10201. [Google Scholar] [CrossRef] [PubMed]
- De Marzo, A.M.; Platz, E.A.; Sutcliffe, S.; Xu, J.; Gronberg, H.; Drake, C.G.; Nakai, Y.; Isaacs, W.B.; Nelson, W.G. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 2007, 7, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; De Marzo, A.M. Prostate cancer and inflammation: The evidence. Histopathology 2012, 60, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Ito, K. Prostate cancer in Asian men. Nat. Rev. Urol. 2014, 11, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High prevalence of mucosa-associated, E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef] [PubMed]
- Dessi, D.; Delogu, G.; Emonte, E.; Catania, M.R.; Fiori, P.L.; Rappelli, P. Long-term survival and intracellular replication of Mycoplasma hominis in Trichomonas vaginalis cells: Potential role of the protozoon in transmitting bacterial infection. Infect. Immun. 2005, 73, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Namiki, K.; Goodison, S.; Porvasnik, S.; Allan, R.W.; Iczkowski, K.A.; Urbanek, C.; Reyes, L.; Sakamoto, N.; Rosser, C.J. Persistent exposure to Mycoplasma induces malignant transformation of human prostate cells. PLoS ONE 2009, 4, e6872. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Chae, S.W.; Kim, H.R.; Chae, H.J. Endoplasmic reticulum stress and cancer. J. Cancer Prev. 2014, 19, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Pluquet, O.; Pourtier, A.; Abbadie, C. The unfolded protein response and cellular senescence. A review in the theme: Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am. J. Physiol. Cell Physiol. 2015, 308, C415–C425. [Google Scholar] [CrossRef] [PubMed]
- Dumartin, L.; Alrawashdeh, W.; Trabulo, S.M.; Radon, T.P.; Steiger, K.; Feakins, R.M.; di Magliano, M.P.; Heeschen, C.; Esposito, I.; Lemoine, N.R.; et al. ER stress protein AGR2 precedes and is involved in the regulation of pancreatic cancer initiation. Oncogene 2016, 36, 3094–3103. [Google Scholar] [CrossRef] [PubMed]
- Celli, J.; Tsolis, R.M. Bacteria, the endoplasmic reticulum and the unfolded protein response: Friends or foes? Nat. Rev. Microbiol. 2015, 13, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kaufman, R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 2014, 14, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Hicks, G.R.; Raikhel, N.V. Protein import into the nucleus: An integrated view. Annu. Rev. Cell Dev. Biol. 1995, 11, 155–188. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.; Ting, C.S.; King, J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001, 11, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Zakariah, M.; Palaniappan, S. Computational prediction of Mycoplasma hominis proteins targeting in nucleus of host cell and their implication in prostate cancer etiology. Tumour Biol. 2016, 37, 10805–10813. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Adapting proteostasis for disease intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kasturi, P.; Bracher, A.; Loew, C.; Zheng, M.; Villella, A.; Garza, D.; Hartl, F.U.; Raychaudhuri, S. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods 2011, 8, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Miao, K.; Li, Y.; Fares, M.; Chen, S.; Zhang, X. A HaloTag-Based Multicolor Fluorogenic Sensor Visualizes and Quantifies Proteome Stress in Live Cells Using Solvatochromic and Molecular Rotor-Based Fluorophores. Biochemistry 2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fares, M.; Dunham, N.P.; Gao, Z.; Miao, K.; Jiang, X.; Bollinger, S.S.; Boal, A.K.; Zhang, X. AgHalo: A Facile Fluorogenic Sensor to Detect Drug-Induced Proteome Stress. Angew. Chem. Int. Ed. Engl. 2018, 56, 8672–8676. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.J.; Ormsby, A.R.; Radwan, M.; Cox, D.; Sharma, A.; Vopel, T.; Ebbinghaus, S.; Oliveberg, M.; Reid, G.E.; Dickson, A.; et al. A biosensor-based framework to measure latent proteostasis capacity. Nat. Commun. 2018, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A. In silico prediction of escherichia coli proteins targeting the host cell nucleus, with special reference to their role in colon cancer etiology. J. Comput. Biol. 2014, 21, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Poutahidis, T.; Cappelle, K.; Levkovich, T.; Lee, C.W.; Doulberis, M.; Ge, Z.; Fox, J.G.; Horwitz, B.H.; Erdman, S.E. Pathogenic intestinal bacteria enhance prostate cancer development via systemic activation of immune cells in mice. PLoS ONE 2013, 8, e73933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karin, M.; Lawrence, T.; Nizet, V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell 2006, 124, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O'Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, Y.; Shen, H.B. Hum-mPLoc 3.0: Prediction enhancement of human protein subcellular localization through modeling the hidden correlations of gene ontology and functional domain features. Bioinformatics 2017, 33, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Raghava, G.P. Prediction of nuclear proteins using SVM and HMM models. BMC Bioinform. 2009, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Entani, T.; Takayama, S.; Tomita, M.; Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 2008, 15, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.B.; Chou, K.C. Hum-mPLoc: An ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem. Biophys. Res. Commun. 2007, 355, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, D191–D198. [Google Scholar]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Pereyre, S.; Sirand-Pugnet, P.; Beven, L.; Charron, A.; Renaudin, H.; Barre, A.; Avenaud, P.; Jacob, D.; Couloux, A.; Barbe, V.; et al. Life on arginine for Mycoplasma hominis: Clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 2009, 5, e1000677. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| NLS | NLS Cutoff | Number of Proteins Targeting ER | Total Number of Proteins in This Range | Percentage |
|---|---|---|---|---|
| Monopartite NLS | 0–3.0 | 15 | 509 | 2.94 |
| 3.0–5.0 | 3 | 32 | 9.3 | |
| 5.0–8.0 | 1 | 16 | 6.25 | |
| >8.0 | 0 | 6 | 0 | |
| Bipartite NLS | 0–3.0 | 6 | 76 | 7.89 |
| 3.0–5.0 | 11 | 290 | 3.79 | |
| 5.0–8.0 | 2 | 194 | 1.03 | |
| >8.0 | 0 | 3 | 0 |
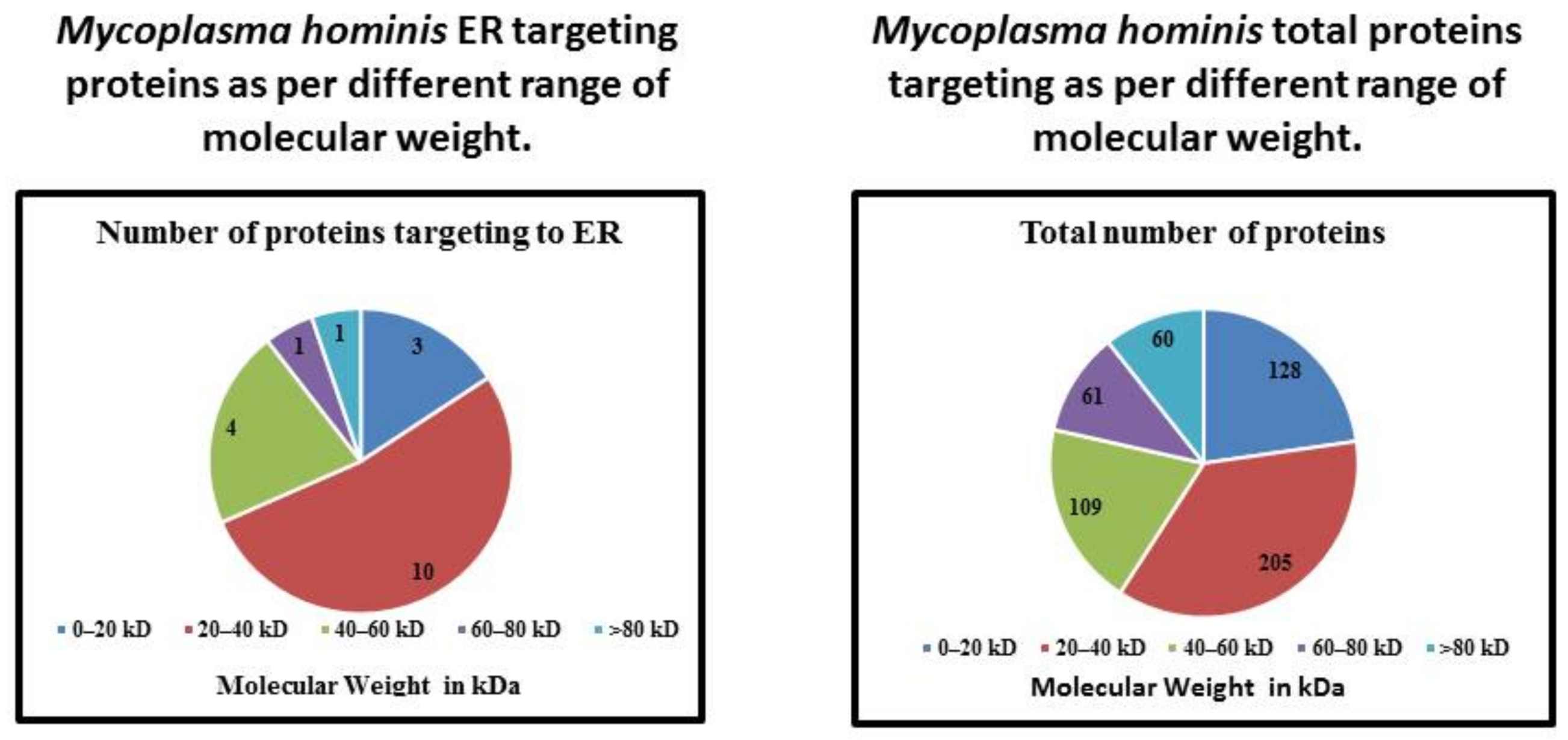
| Molecular Weight | Number of Proteins Targeting to ER | Total Number of Proteins | Percentage |
|---|---|---|---|
| 0–20 kDa | 3 | 128 | 2.34 |
| 20–40 kDa | 10 | 205 | 4.87 |
| 40–60 kDa | 4 | 109 | 3.66 |
| 60–80 kDa | 1 | 61 | 1.63 |
| >80 kDa | 1 | 60 | 1.66 |
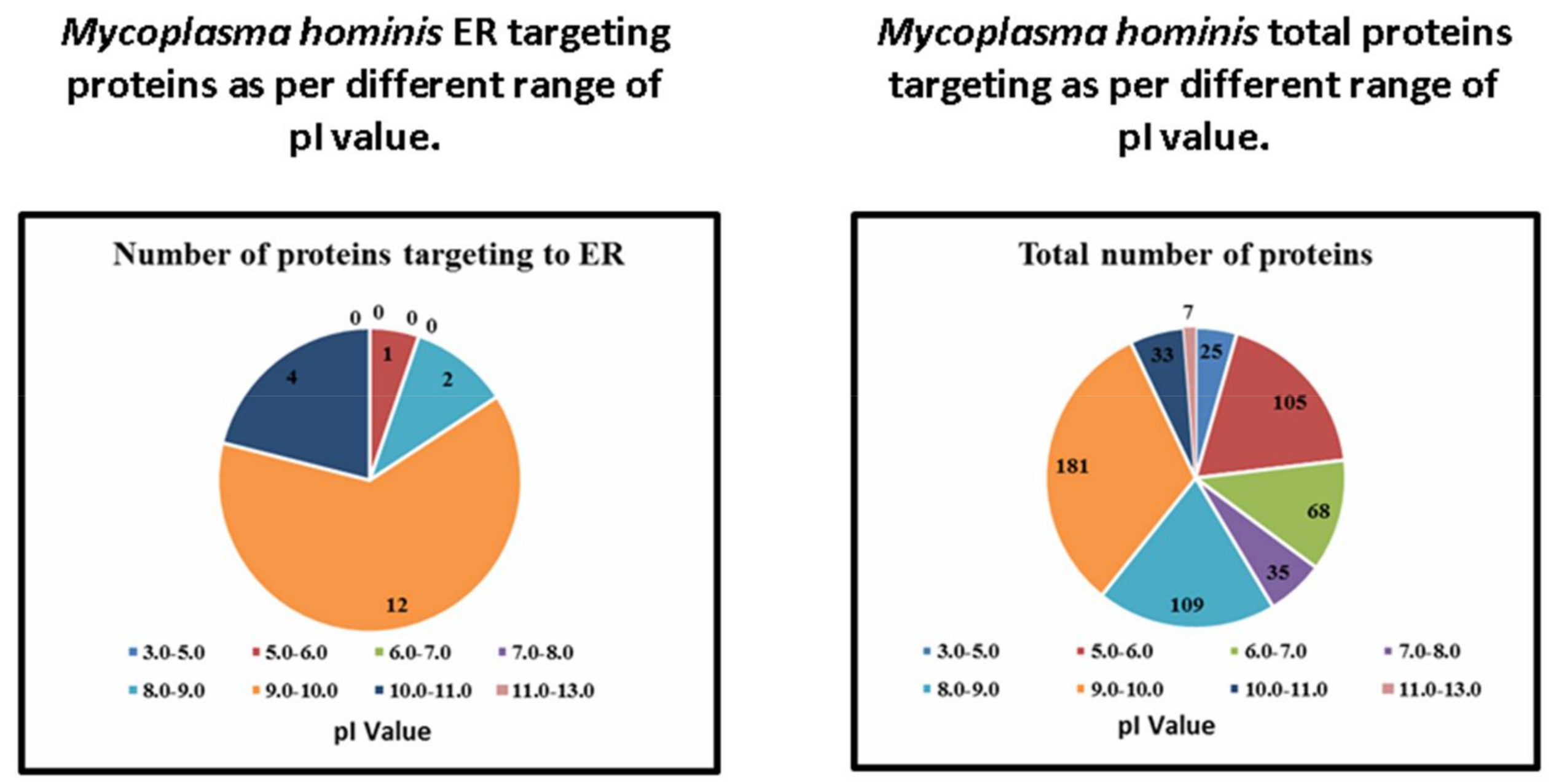
| Range of pI Value | Number of Proteins Targeting to ER | Total Number of Proteins | Percentage |
|---|---|---|---|
| 3.0–5.0 | 0 | 25 | 0 |
| 5.0–6.0 | 1 | 105 | 0.95 |
| 6.0–7.0 | 0 | 68 | 0 |
| 7.0–8.0 | 0 | 35 | 0 |
| 8.0–9.0 | 2 | 109 | 1.83 |
| 9.0–10.0 | 12 | 181 | 6.62 |
| 10.0–11.0 | 4 | 33 | 12.12 |
| 12.0–13.0 | 0 | 7 | 0 |
| Accession Number | Protein Name | Function in Bacteria | Protein Existence | pI | Molecular Weight | NLS Mapper | Hum-mPLoc 3.0 | |
|---|---|---|---|---|---|---|---|---|
| Monopartite | Bipartite | |||||||
| A0A097NT54 | Uncharacterized protein | Unknown | Protein predicted | 10.17 | 13,450 | 0 | 22 | Endoplasmic reticulum |
| A0A097NSU8 | Uncharacterized protein | Unknown | Protein predicted | 5.45 | 36,963 | 0 | 3.8 | Endoplasmic reticulum |
| A0A097NTC6 | Lipoprotein signal peptidase (EC 3.4.23.36) | Aspartic-type endopeptidase activity. | Protein inferred from homology | 8.44 | 23,620 | 0 | 3.3 | Endoplasmic reticulum |
| A0A097NTZ8 | Cation transporting P-type ATPase (EC 3.6.3.8) | ATP binding, calcium-transporting ATPase activity, and metal ion binding activity. | Protein inferred from homology | 8.56 | 107,148 | 2 | 6.1 | Endoplasmic reticulum |
| A0A097NT87 | Protein translocase subunit SecY | Intracellular protein transmembrane transport activity, protein transport activity hrough the Sec complex | Protein inferred from homology | 9.51 | 54,926 | 6 | 3.9 | Endoplasmic reticulum |
| A0A097NTV1 | Membrane protein | Unknown | Protein predicted | 9.54 | 25,294 | 0 | 5.5 | Endoplasmic reticulum |
| A0A097NTF0 | Prolipoprotein diacylglyceryl transferase (EC 2.4.99.-) | Phosphatidylglycerol-prolipoprotein diacylglyceryl transferase activity and involved in lipoprotein biosynthetic process. | Protein inferred from homology | 9.56 | 35,944 | 0 | 3.8 | Endoplasmic reticulum |
| A0A097NSJ5 | Potassium transporter KtrB | Cation transmembrane transporter activity. | Protein predicted | 9.6 | 58,434 | 0 | 3 | Endoplasmic reticulum |
| A0A097NT10 | ComEC/Rec2-related protein | Unknown | Protein predicted | 9.65 | 54,059 | 0 | 4.4 | Endoplasmic reticulum |
| A0A097NSI4 | Uncharacterized protein | Unknown | Protein predicted | 9.66 | 38,283 | 0 | 3.3 | Endoplasmic reticulum |
| A0A097NSM2 | Membrane protein | Unknown | Protein predicted | 9.72 | 33,519 | 0 | 3.4 | Endoplasmic reticulum |
| A0A097NSJ8 | 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) | 1-acylglycerol-3-phosphate O-acyltransferase activity. | Protein predicted | 9.79 | 31,034 | 0 | 4.7 | Endoplasmic reticulum |
| A0A097NT14 | Cobalt ABC transporter permease | Unknown | Protein predicted | 9.85 | 35,998 | 5 | 4.4 | Endoplasmic reticulum |
| A0A097NTJ8 | ABC transporter permease | Transporter activity. | Protein inferred from homology | 9.86 | 65,590 | 0 | 2.9 | Endoplasmic reticulum |
| A0A097NTE8 | Uncharacterized protein | Unknown | Protein predicted | 9.9 | 57,701 | 0 | 5 | Endoplasmic reticulum |
| A0A097NSN9 | Uncharacterized protein | Unknown | Protein predicted | 9.97 | 14,493 | 0 | 2.6 | Endoplasmic reticulum |
| A0A097NSG1 | Uncharacterized protein | Unknown | Protein predicted | 10.1 | 17,661 | 4 | 2.6 | Endoplasmic reticulum |
| A0A097NTR2 | Membrane protein | Transporter activity. | Protein predicted | 10.19 | 33,456 | 4 | 3.2 | Endoplasmic reticulum |
| A0A097NT45 | Membrane protein | Unknown | Protein predicted | 10.47 | 20,769 | 0 | 2.5 | Endoplasmic reticulum |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakariah, M.; Khan, S.; Chaudhary, A.A.; Rolfo, C.; Ben Ismail, M.M.; Alotaibi, Y.A. To Decipher the Mycoplasma hominis Proteins Targeting into the Endoplasmic Reticulum and Their Implications in Prostate Cancer Etiology Using Next-Generation Sequencing Data. Molecules 2018, 23, 994. https://doi.org/10.3390/molecules23050994
Zakariah M, Khan S, Chaudhary AA, Rolfo C, Ben Ismail MM, Alotaibi YA. To Decipher the Mycoplasma hominis Proteins Targeting into the Endoplasmic Reticulum and Their Implications in Prostate Cancer Etiology Using Next-Generation Sequencing Data. Molecules. 2018; 23(5):994. https://doi.org/10.3390/molecules23050994
Chicago/Turabian StyleZakariah, Mohammed, Shahanavaj Khan, Anis Ahmad Chaudhary, Christian Rolfo, Mohamed Maher Ben Ismail, and Yousef Ajami Alotaibi. 2018. "To Decipher the Mycoplasma hominis Proteins Targeting into the Endoplasmic Reticulum and Their Implications in Prostate Cancer Etiology Using Next-Generation Sequencing Data" Molecules 23, no. 5: 994. https://doi.org/10.3390/molecules23050994






