Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Mechanisms of the HIV-1 Drug Resistance to Reverse Transcriptase Inhibitors
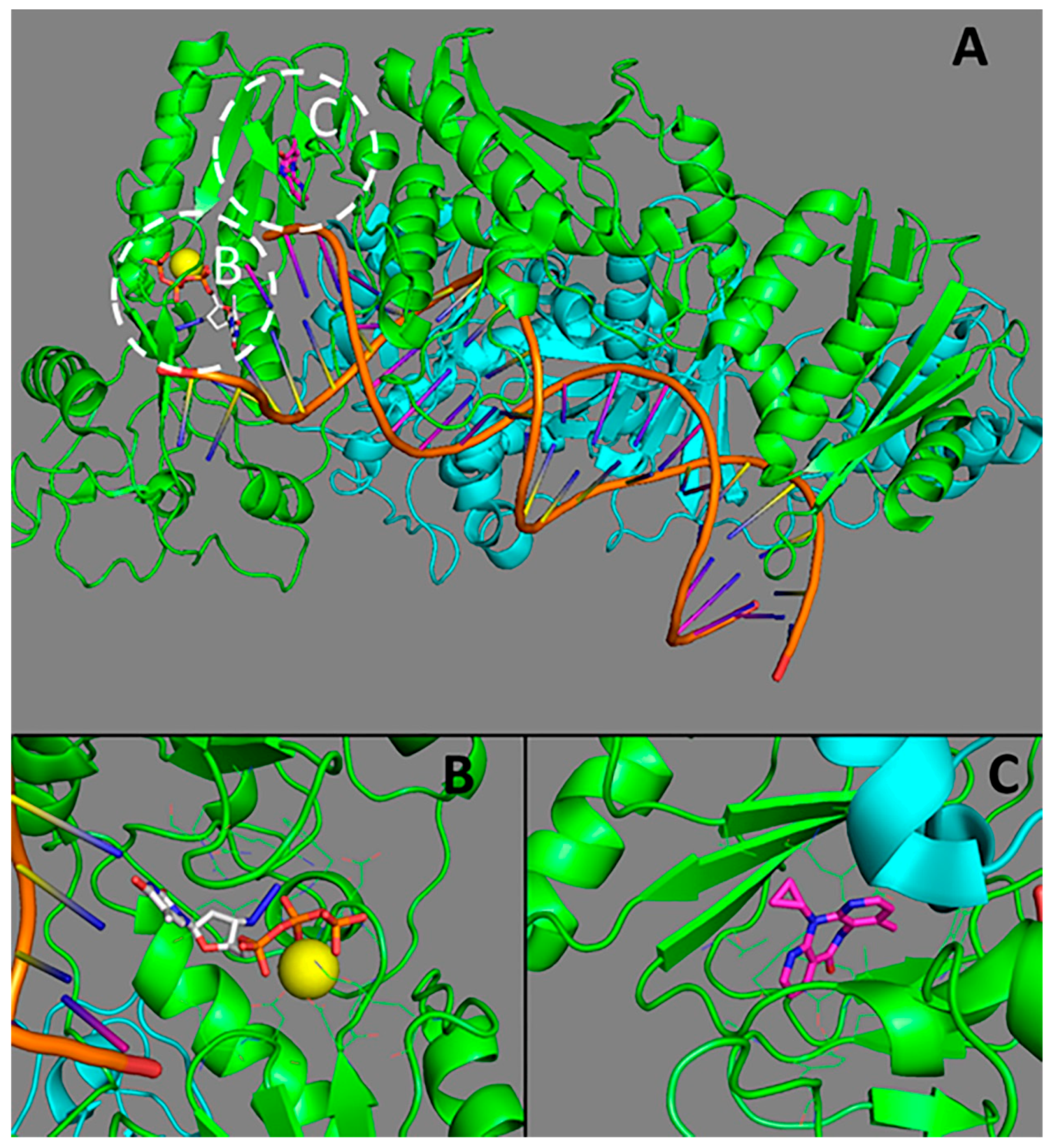
2.1.1. Three-Dimensional Structure of HIV-1 Reverse Transcriptase
2.1.2. Mechanisms of Reverse Transcriptase Inhibition
2.2. Molecular Docking in Studies of HIV-1 Drug Resistance to Reverse Transcriptase Inhibitors
2.2.1. Data on Three-Dimensional Complexes
2.2.2. Applications of Molecular Docking for the Development of New HIV Reverse Transcriptase Inhibitors
2.3. Perspectives of Molecular Docking in the Studies of HIV-1 Resistance
3. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Connor, E.M.; Sperling, R.S.; Gelber, R.; Kiselev, P.; Scott, G.; O’Sullivan, M.J.; VanDyke, R.; Bey, M.; Shearer, W.; Jacobson, R.L. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N. Engl. J. Med. 1994, 331, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.H.; Venkatesh, K.K. Antiretroviral therapy as HIV prevention: Status and prospects. Am. J. Public Health 2010, 100, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- McMahon, J.M.; Myers, J.E.; Kurth, A.E.; Cohen, S.E.; Mannheimer, S.B.; Simmons, J.; Pouget, E.R.; Trabold, N.; Haberer, J.E. Oral Pre-Exposure Prophylaxis (PrEP) for Prevention of HIV in Serodiscordant Heterosexual Couples in the United States: Opportunities and Challenges. AIDS Patient Care STDs 2014, 28, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Mugwanya, K.K.; Baeten, J.M. Safety of oral tenofovir disoproxil fumarate-based pre-exposure prophylaxis for HIV prevention. Expert Opin. Drug Saf. 2016, 15, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Buckheit, R.W.; Watson, K.M.; Morrow, K.M.; Ham, A.S. Development of topical microbicides to prevent the sexual transmission of HIV. Antivir. Res. 2010, 85, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Ham, A.S.; Nugent, S.T.; Peters, J.J.; Katz, D.F.; Shelter, C.M.; Dezzutti, C.S.; Boczar, A.D.; Buckheit, K.W.; Buckheit, R.W. The rational design and development of a dual chamber vaginal/rectal microbicide gel formulation for HIV prevention. Antivir. Res. 2015, 120, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Guasch, L.; Zakharov, A.V.; Tarasova, O.A.; Poroikov, V.V.; Liao, C.; Nicklaus, M.C. Novel HIV-1 Integrase Inhibitor Development by Virtual Screening Based on QSAR Models. Curr. Top. Med. Chem. 2016, 16, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.; Eleftheriou, P.; Poroikov, V. Anti-HIV Agents: Current Status and Recent Trends; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Santos, L.H.; Ferreira, R.S.; Caffarena, E.R. Computational drug design strategies applied to the modelling of human immunodeficiency virus-1 reverse transcriptase inhibitors. Mem. Inst. Oswaldo Cruz 2015, 110, 847–864. [Google Scholar] [CrossRef] [PubMed]
- Dharmalingam, T.; Udhaya, V.; Umaarasu, T.; Elangovan, V.; Rajesh, S.V.; Shanmugam, G. Prediction of drug-resistance using genotypic and docking analysis among anti-retroviral therapy naïve and first-line treatment failures in Salem, Tamil Nadu, India. Curr. HIV Res. 2015, 13, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Distinto, S.; Liedl, K.R.; Markt, P.; Rollinger, J.M.; Schuster, D.; Spitzer, G.M.; Wolber, G. Development of anti-viral agents using molecular modeling and virtual screening techniques. Infect. Disord. Drug Targets 2011, 11, 64–93. [Google Scholar] [CrossRef] [PubMed]
- Frey, K.M. Structure-enhanced methods in the development of non-nucleoside inhibitors targeting HIV reverse transcriptase variants. Future Microbiol. 2015, 10, 1767–1772. [Google Scholar] [CrossRef] [PubMed]
- Vanangamudi, M.; Poongavanam, V.; Namasivayam, V. HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors: SAR and Lead Optimization Using CoMFA and CoMSIA Studies (1995–2016). Curr. Med. Chem. 2017, 24, 3774–3812. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Smerdon, S.J.; Jäger, J.; Kohlstaedt, L.A.; Rice, P.A.; Friedman, J.M.; Steitz, T.A. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. USA 1994, 91, 7242–7246. [Google Scholar] [CrossRef] [PubMed]
- London, R.E. Structural Maturation of HIV-1 Reverse Transcriptase-A Metamorphic Solution to Genomic Instability. Viruses 2016, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Boso, G.; Örvell, C.; Somia, N.V. The nature of the N-terminal amino acid residue of HIV-1 RNase H is critical for the stability of reverse transcriptase in viral particles. J. Virol. 2015, 89, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Wapling, J.; Moore, K.L.; Sonza, S.; Mak, J.; Tachedjian, G. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J. Virol. 2005, 79, 10247–10257. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Berducou, C.; Mery, J.; Heitz, F.; Divita, G. The Thumb Domain of the P51-Subunit Is Essential for Activation of HIV Reverse Transcriptase. Biochemistry 1999, 38, 15097–15103. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Miller, J.T.; Lapkouski, M.; Tian, L.; Yang, W.; Le Grice, S.F.J. Examining the role of the HIV-1 reverse transcriptase p51 subunit in positioning and hydrolysis of RNA/DNA hybrids. J. Biol. Chem. 2013, 288, 16177–16184. [Google Scholar] [CrossRef] [PubMed]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [PubMed]
- Kaushik, N.; Rege, N.; Yadav, P.N.; Sarafianos, S.G.; Modak, M.J.; Pandey, V.N. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry 1996, 35, 11536–11546. [Google Scholar] [CrossRef] [PubMed]
- Rakik, A.; Ait-Khaled, M.; Griffin, P.; Thomas, T.A.; Tisdale, M.; Kleim, J.P. A novel genotype encoding a single amino acid insertion and five other substitutions between residues 64 and 74 of the HIV-1 reverse transcriptase confers high-level cross-resistance to nucleoside reverse transcriptase inhibitors. Abacavir CNA2007 International Study Group. J. Acquir. Immune Defic. Syndr. 1999, 22, 139–145. [Google Scholar] [PubMed]
- Garforth, S.J.; Lwatula, C.; Prasad, V.R. The lysine 65 residue in HIV-1 reverse transcriptase function and in nucleoside analog drug resistance. Viruses 2014, 6, 4080–4094. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.L.; Sarafianos, S.G.; Arnold, E.; Hughes, S.H. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 2000, 97, 3056–3061. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Martinez, S.E.; Arnold, E. Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-S.; Hughes, S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. D Struct. Biol. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Das, K.; Martinez, S.E.; Bauman, J.D.; Arnold, E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 2012, 19, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Martinez, S.E.; Bandwar, R.P.; Arnold, E. Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: Insights into requirements for RNase H cleavage. Nucleic Acids Res. 2014, 42, 8125–8137. [Google Scholar] [CrossRef] [PubMed]
- Iyidogan, P.; Anderson, K. Current Perspectives on HIV-1 Antiretroviral Drug Resistance. Viruses 2014, 6, 4095–4139. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Clark, A.D.; Ding, J.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 1999, 96, 10027–10032. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Hughes, S.H.; Arnold, E. Designing anti-AIDS drugs targeting the major mechanism of HIV-1 RT resistance to nucleoside analog drugs. Int. J. Biochem. Cell Biol. 2004, 36, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Deval, J.; Courcambeck, J.; Selmi, B.; Boretto, J.; Canard, B. Structural determinants and molecular mechanisms for the resistance of HIV-1 RT to nucleoside analogues. Curr. Drug Metab. 2004, 5, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Miranda, L.R.; Götte, M.; Liang, F.; Kuritzkes, D.R. The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations. Antimicrob. Agents Chemother. 2005, 49, 2648–2656. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Bandwar, R.P.; White, K.L.; Feng, J.Y.; Sarafianos, S.G.; Tuske, S.; Tu, X.; Clark, A.D.; Boyer, P.L.; Hou, X.; et al. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J. Biol. Chem. 2009, 284, 35092–35100. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antivir. Res. 2013, 98, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.-Y.; Sankaran, K.; Varghese, V.; Winters, M.A.; Hurt, C.B.; Eron, J.J.; Parkin, N.; Holmes, S.P.; Holodniy, M.; Shafer, R.W. HIV-1 Protease, Reverse Transcriptase, and Integrase Variation. J. Virol. 2016, 90, 6058–6070. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Coutsinos, D. The K65R mutation in HIV-1 reverse transcriptase: Genetic barriers, resistance profile and clinical implications. HIV Ther. 2009, 3, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Scarth, B.J.; White, K.L.; Chen, J.M.; Lansdon, E.B.; Swaminathan, S.; Miller, M.D.; Götte, M. Mechanism of Resistance to GS-9148 Conferred by the Q151L Mutation in HIV-1 Reverse Transcriptase. Antimicrob. Agents Chemother. 2011, 55, 2662–2669. [Google Scholar] [CrossRef] [PubMed]
- Merluzzi, V.J.; Hargrave, K.D.; Labadia, M.; Grozinger, K.; Skoog, M.; Wu, J.C.; Shih, C.K.; Eckner, K.; Hattox, S.; Adams, J. Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science 1990, 250, 1411–1413. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Das, K.; Hsiou, Y.; Sarafianos, S.G.; Clark, A.D.; Jacobo-Molina, A.; Tantillo, C.; Hughes, S.H.; Arnold, E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J. Mol. Biol. 1998, 284, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.-T.; Munshi, V.; Lu, M.; Feng, M.; Hrin-Solt, R.; McKenna, P.; Hazuda, D.; Miller, M. Mechanistic Study of Common Non-Nucleoside Reverse Transcriptase Inhibitor-Resistant Mutations with K103N and Y181C Substitutions. Viruses 2016, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Kroeger Smith, M.B.; Michejda, C.J.; Hughes, S.H.; Boyer, P.L.; Janssen, P.A.; Andries, K.; Buckheit, R.W.; Smith, R.H. Molecular modeling of HIV-1 reverse transcriptase drug-resistant mutant strains: Implications for the mechanism of polymerase action. Protein Eng. 1997, 10, 1379–1383. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Tantillo, C.; Clark, A.D.; Ding, J.; Whitcomb, J.M.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001, 20, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, C.; Wisniewski, M.; Jacques, P.S.; Le Grice, S.F.; Fay, P.J.; Bambara, R.A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 1997, 272, 11157–11164. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami, M.; Götte, M. Effects of mutations in the connection and RNase H domains of HIV-1 reverse transcriptase on drug susceptibility. AIDS Rev. 2008, 10, 224–235. [Google Scholar] [PubMed]
- Kharytonchyk, S.; King, S.R.; Ndongmo, C.B.; Stilger, K.L.; An, W.; Telesnitsky, A. Resolution of Specific Nucleotide Mismatches by Wild-Type and AZT-Resistant Reverse Transcriptases during HIV-1 Replication. J. Mol. Biol. 2016, 428, 2275–2288. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.P.; Moore, C.L.; Gotuzzo, E.; Nwizu, C.; Kamarulzaman, A.; Chetchotisakd, P.; van Wyk, J.; Teppler, H.; Kumarasamy, N.; Molina, J.-M.; et al. Antiretroviral Resistance after First-Line Antiretroviral Therapy Failure in Diverse HIV-1 Subtypes in the SECOND-LINE Study. AIDS Res. Hum. Retrovir. 2016, 32, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, N.M.; Gesto, D.; Oliveira, E.F.; Santos-Martins, D.; Brás, N.F.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Receptor-based virtual screening protocol for drug discovery. Arch. Biochem. Biophys. 2015, 582, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Caflisch, A. Protein structure-based drug design: From docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018, 48, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Shoichet, B.K.; McGovern, S.L.; Wei, B.; Irwin, J.J. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002, 6, 439–446. [Google Scholar] [CrossRef]
- Kroemer, R.T. Structure-based drug design: Docking and scoring. Curr. Protein Pept. Sci. 2007, 8, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Springer, C.; Adalsteinsson, H.; Young, M.M.; Kegelmeyer, P.W.; Roe, D.C. PostDOCK: A structural, empirical approach to scoring protein ligand complexes. J. Med. Chem. 2005, 48, 6821–6831. [Google Scholar] [CrossRef] [PubMed]
- Perola, E.; Walters, W.P.; Charifson, P.S. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins Struct. Funct. Bioinform. 2004, 56, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Kalyanaraman, C.; Irwin, J.J.; Jacobson, M.P. Physics-Based Scoring of Protein−Ligand Complexes: Enrichment of Known Inhibitors in Large-Scale Virtual Screening. J. Chem. Inf. Model. 2006, 46, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Kalyanaraman, C.; Bernacki, K.; Jacobson, M.P. Molecular mechanics methods for predicting protein–ligand binding. Phys. Chem. Chem. Phys. 2006, 8, 5166–5177. [Google Scholar] [CrossRef] [PubMed]
- Floriano, W.B.; Vaidehi, N.; Zamanakos, G.; Goddard, W.A. HierVLS Hierarchical Docking Protocol for Virtual Ligand Screening of Large-Molecule Databases. J. Med. Chem. 2004, 47, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Miteva, M.A.; Lee, W.H.; Montes, M.O.; Villoutreix, B.O. Fast Structure-Based Virtual Ligand Screening Combining FRED, DOCK, and Surflex. J. Med. Chem. 2005, 48, 6012–6022. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput.-Aided Mol. Des. 2007, 21, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Gulnik, S.; Everitt, L.; Kaplan, A.H. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 2003, 77, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Visseaux, B.; Armenia, D.; Morand-Joubert, L.; Artese, A.; Charpentier, C.; Van Den Eede, P.; Costa, G.; Alcaro, S.; Wirden, M.; et al. Identification of a rare mutation at reverse transcriptase Lys65 (K65E) in HIV-1-infected patients failing on nucleos(t)ide reverse transcriptase inhibitors. J. Antimicrob. Chemother. 2013, 68, 2199–2204. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.; Surdo, M.; Di Maio, V.C.; Di Santo, F.; Costa, G.; Parrotta, L.; Romeo, I.; Gori, C.; Santoro, M.M.; Fedele, V.; et al. The HIV-1 reverse transcriptase polymorphism A98S improves the response to tenofovir disoproxil fumarate+emtricitabine-containing HAART both in vivo and in vitro. J. Glob. Antimicrob. Resist. 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, W.; Kang, D.; Lu, X.; Li, X.; Liu, Z.; Huang, B.; Daelemans, D.; Pannecouque, C.; De Clercq, E.; et al. Design, synthesis and anti-HIV evaluation of novel diarylpyridine derivatives targeting the entrance channel of NNRTI binding pocket. Eur. J. Med. Chem. 2016, 109, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Yin, H.; De Clercq, E.; Pannecouque, C.; Meng, G.; Chen, F. Discovery of biphenyl-substituted diarylpyrimidines as non-nucleoside reverse transcriptase inhibitors with high potency against wild-type and mutant HIV-1. Eur. J. Med. Chem. 2018, 145, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, L.; Zhang, X.; Lau, T.C.K.; Tsui, S.K.W.; Kang, Y.; Zheng, P.; Zheng, B.; Liu, G.; Chen, Z. F18, a novel small-molecule nonnucleoside reverse transcriptase inhibitor, inhibits HIV-1 replication using distinct binding motifs as demonstrated by resistance selection and docking analysis. Antimicrob. Agents Chemother. 2012, 56, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Samanta, P.N.; Das, K.K. Inhibition activities of catechol diether based non-nucleoside inhibitors against the HIV reverse transcriptase variants: Insights from molecular docking and ONIOM calculations. J. Mol. Graph. Model. 2017, 75, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Pagano, N.; Teriete, P.; Mattmann, M.E.; Yang, L.; Snyder, B.A.; Cai, Z.; Heil, M.L.; Cosford, N.D.P. An integrated chemical biology approach reveals the mechanism of action of HIV replication inhibitors. Bioorg. Med. Chem. 2017, 25, 6248–6265. [Google Scholar] [CrossRef] [PubMed]
- Miceli, L.; Teixeira, V.; Castro, H.; Rodrigues, C.; Mello, J.; Albuquerque, M.; Cabral, L.; de Brito, M.; de Souza, A. Molecular Docking Studies of Marine Diterpenes as Inhibitors of Wild-Type and Mutants HIV-1 Reverse Transcriptase. Mar. Drugs 2013, 11, 4127–4143. [Google Scholar] [CrossRef] [PubMed]
- Nizami, B.; Sydow, D.; Wolber, G.; Honarparvar, B. Molecular insight on the binding of NNRTI to K103N mutated HIV-1 RT: Molecular dynamics simulations and dynamic pharmacophore analysis. Mol. BioSyst. 2016, 12, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and function of HIV-1 reverse transcriptase: Molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [PubMed]
- Derudas, M.; Vanpouille, C.; Carta, D.; Zicari, S.; Andrei, G.; Snoeck, R.; Brancale, A.; Margolis, L.; Balzarini, J.; McGuigan, C. Virtual Screening of Acyclovir Derivatives as Potential Antiviral Agents: Design, Synthesis, and Biological Evaluation of New Acyclic Nucleoside ProTides. J. Med. Chem. 2017, 60, 7876–7896. [Google Scholar] [CrossRef] [PubMed]
- Ferro, S.; Buemi, M.R.; De Luca, L.; Agharbaoui, F.E.; Pannecouque, C.; Monforte, A.-M. Searching for novel N1-substituted benzimidazol-2-ones as non-nucleoside HIV-1 RT inhibitors. Bioorg. Med. Chem. 2017, 25, 3861–3870. [Google Scholar] [CrossRef] [PubMed]
- Suryawanshi, R.; Jadhav, S.; Makwana, N.; Desai, D.; Chaturbhuj, D.; Sonawani, A.; Idicula-Thomas, S.; Murugesan, V.; Katti, S.B.; Tripathy, S.; et al. Evaluation of 4-thiazolidinone derivatives as potential reverse transcriptase inhibitors against HIV-1 drug resistant strains. Bioorg. Chem. 2017, 71, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Devale, T.L.; Parikh, J.; Miniyar, P.; Sharma, P.; Shrivastava, B.; Murumkar, P. Dihydropyrimidinone-isatin hybrids as novel non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg. Chem. 2017, 70, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Chander, S.; Wang, P.; Ashok, P.; Yang, L.-M.; Zheng, Y.-T.; Sankaranarayanan, M. Design, synthesis and anti-HIV-1 RT evaluation of 2-(benzyl(4-chlorophenyl)amino)-1-(piperazin-1-yl)ethanone derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Ivetac, A.; Swift, S.E.; Boyer, P.L.; Diaz, A.; Naughton, J.; Young, J.A.T.; Hughes, S.H.; McCammon, J.A. Discovery of novel inhibitors of HIV-1 reverse transcriptase through virtual screening of experimental and theoretical ensembles. Chem. Biol. Drug Des. 2014, 83, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Frączek, T.; Siwek, A.; Paneth, P. Assessing molecular docking tools for relative biological activity prediction: A case study of triazole HIV-1 NNRTIs. J. Chem. Inf. Model. 2013, 53, 3326–3342. [Google Scholar] [CrossRef] [PubMed]
- Vadivelan, S.; Deeksha, T.N.; Arun, S.; Machiraju, P.K.; Gundla, R.; Sinha, B.N.; Jagarlapudi, S.A.R.P. Virtual screening studies on HIV-1 reverse transcriptase inhibitors to design potent leads. Eur. J. Med. Chem. 2011, 46, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Tintori, C.; Corona, A.; Esposito, F.; Brai, A.; Grandi, N.; Ceresola, E.R.; Clementi, M.; Canducci, F.; Tramontano, E.; Botta, M. Inhibition of HIV-1 Reverse Transcriptase Dimerization by Small Molecules. ChemBioChem 2016, 17, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- García-Godoy, M.; López-Camacho, E.; García-Nieto, J.; Nebro, A.; Aldana-Montes, J. Solving Molecular Docking Problems with Multi-Objective Metaheuristics. Molecules 2015, 20, 10154–10183. [Google Scholar] [CrossRef] [PubMed]
- Awuni, Y.; Mu, Y. Reduction of False Positives in Structure-Based Virtual Screening When Receptor Plasticity Is Considered. Molecules 2015, 20, 5152–5164. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Chermak, E.; Cavallo, L. Analysis and Ranking of Protein-Protein Docking Models Using Inter-Residue Contacts and Inter-Molecular Contact Maps. Molecules 2015, 20, 12045–12060. [Google Scholar] [CrossRef] [PubMed]
- Kumalo, H.; Bhakat, S.; Soliman, M. Theory and Applications of Covalent Docking in Drug Discovery: Merits and Pitfalls. Molecules 2015, 20, 1984–2000. [Google Scholar] [CrossRef] [PubMed]
- Atkovska, K.; Samsonov, S.A.; Paszkowski-Rogacz, M.; Pisabarro, M.T. Multipose binding in molecular docking. Int. J. Mol. Sci. 2014, 15, 2622–2645. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Y. Improving scoring-docking-screening powers of protein-ligand scoring functions using random forest. J. Comput. Chem. 2017, 38, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kinnings, S.L.; Liu, N.; Tonge, P.J.; Jackson, R.M.; Xie, L.; Bourne, P.E. A Machine Learning-Based Method to Improve Docking Scoring Functions and Its Application to Drug Repurposing. J. Chem. Inf. Model. 2011, 51, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.U.; Aleksandrova, A.; Roessler, F.D.; Ballester, P.J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2015, 5, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Ragoza, M.; Hochuli, J.; Idrobo, E.; Sunseri, J.; Koes, D.R. Protein–Ligand Scoring with Convolutional Neural Networks. J. Chem. Inf. Model. 2017, 57, 942–957. [Google Scholar] [CrossRef] [PubMed]
| Drug Name | Class | The Major Positions and Mutations Leading to Resistance |
|---|---|---|
| Lamivudine | NRTI 1 | K65R, M184V/I |
| Emtricitabine | NRTI | K65R, M184V/I |
| Abacavir | NRTI | K65R, K70I, L74V/I, Y115F, M184V/I |
| Tenofovir | NRTI | M41L, K65R, K70R, T215F/Y |
| Zidovudine | NRTI | M41L, D67N, K70R, L210W, T215F/Y, K219Q/E |
| Nevirapine | NNRTI 2 | L100I, K101E/P, K103N/S, V106A/M, Y181C/I/V, Y188L/C/H, G190A/S/E/Q, M230L |
| Efavirenz | NNRTI | K103N/S, V106A/M, Y181C/I/V, Y188L/C/H, G190A/S/E/Q, M230L |
| Etravirine | NNRTI | L100I, K101E/P, Y181C/I/V, Y188L/C/H, G190A/S/E/Q, M230L |
| Rilpivirine | NNRTI | L100I, K101E/P, Y181C/I/V, Y188L/C/H, G190A/S/E/Q, M230L |
| Mutation/Wild-Type Protein | Number of Complexes | PDB IDs |
|---|---|---|
| Wild-type | 120 | 1C0T; 1C0U; 1DLO; 1DTQ; 1DTT; 1FK9; 1HMV; 1HNI; 1HNV; 1HVU; 1HYS; 1IKW; 1JLQ; 1KLM; 1N5Y; 1N6Q, 1R0A; 1REV; 1RT1; 1RT2; 1RT4; 1RT5; 1RT6; 1RT7; 1RTD; 1RTH; 1RTI; 1RTJ; 1S6P; 1S6Q; 1S9E; 1S9G; 1SUQ; 1T03; 1T05; 1TKT; 1TKX; 1TKZ; 1TL1; 1TL3; 1TV6; 1TVR; 1VRT; 1VRU; 2B5J; 2B6A; 2BAN; 2BE2; 2HMI; 2HND; 2I5J; 2JLE; 2OPP; 2RF2; 2RKI; 2VG5; 2VG6; 2VG7; 2WON; 2YKM; 2YKN; 2YNG; 2YNH; 2YNI; 2ZD1; 3C6T; 3C6U; 3DI6; 3DLE; 3DLG; 3DRP; 3DYA; 3E01; 3FFI; 3HVT; 3I0R; 3I0S; 3IG1; 3IRX; 3IS9; 3ISN; 3ITH; 3KJV; 3KK1; 3KK2; 3KK3; 3KLF; 3LAK; 3LAL; 3LAM; 3LAN; 3LP0; 3LP1; 3LP2; 3M8P; 3M8Q; 3MEC; 3MEE; 3NBP; 3QIP; 3T19; 3V4I; 3V6D; 3V81; 4B3O; 4B3P; 4B3Q; 4I7F; 4ID5; 4KV8; 4LSL; 4NCG; 4PQU; 4PUO; 4PWD; 4Q0B; 4R5P; 5CYM; 5HLF; 1BQM |
| Y188C | 4 | 1JLE; 1JLG; 2OPS; 2YNF |
| K103N | 18 | 1FKO; 1FKP; 1HPZ; 1HQE; 1HQU; 1IKV; 1IKX; 1IKY; 1SV5; 2WOM; 3DOK; 3DM2; 3DRS; 3MED; 3MEG; 3T1A; 3TAM; 1SV5 |
| K103N/Y181C | 7 | 2IAJ; 2IC3; 3BGR; 3DM2; 4RW4; 4RW7; 5FDL |
| L100I/K103N | 1 | 2ZE2 |
| Y181C | 9 | 1C1B; 1C1C; 1JKH; 1JLA; 1JLB; 1JLC; 1UWB; 3DRR; 4RW6 |
| Y188C | 4 | 1JLE; 1JLF; 1JLG; 2OPS |
| Y188L | 2 | 1BQN; 2YNF |
| Other mutations or combinations | 53 | 1EET; 1EP4; 1J5O; 1LW0; 1LW2; 1LWC; 1LWE; 1LWF; 1QE1; 1RT3; 1S1T; 1S1U; 1S1V; 1S1W; 1S1X; 2HNY; 2HNZ; 2OPQ; 2OPR; 2OPS; 2ZE2; 3DMJ; 3DOL; 3JSM; 3JYT; 3KLE; 3KLG; 3KLH; 3KLI; 3MED; 3QLH; 3QO9; 4DG1; 4G1Q; 4H4M; 4H4O; 4I2P; 4I2Q; 4ICL; 4IDK; 4IFV; 4IFY; 4IG0; 4IG3; 4KFB; 4KKO; 4KO0; 4KSE; 4LS; 4MFB; 4O44; 4O4G; 4RW8; 4RW9; 4WE1; 4ZHR; 5C24; 5C25; 5C42; 5CYQ; 5HBM |
| Mutati-Ons | PDB ID | Mutant RT Preparation | Class of Drug or Chemical | Docking Algorithm, Package | Energy Minimization Algorithm in Post-Docking Protocol | Reference |
|---|---|---|---|---|---|---|
| K65E | 1RTD | Computationally MacroModel 2.0 | NRTIs, marketed drugs | Autodock 4.0 | AMBER (ligand) | [65] |
| A98S | Computationally | NRTIs, marketed drugs | - | N/D * | [66] | |
| WT K103N | 3MED | PDB complexes | NNRTIs diaryl-pyridine derivatives (DAPYs) | Surflex-Dock (Sybyl-X 2.0) | N/D | [67] |
| V106A and F227L | 2ZD1 | Computationally | NNRTIs diaryl-pyrimidine analogs | Sybyl-X 1.2 | N/D | [68] |
| WT L100I Y181C | 1VRT 1S1U 1JLB | Experimental (PDB) | NNRTIs (+)-calanoide analog | Autodock 4.2 | AMBER (ligand-protein energy calculation) Conformation clusterization using RMSD | [69] |
| WT Y181C | 4H4M 4RW6 4RW4 4RW8 4RW9 4RW7 | Computationally using PDB complexes 4RW6, 4RW4, 4H4M | NNRTIs catechol diether based NNRTIs | Autodock 4.2Autodock Vina ArgusLab 4.0.1 | ONIOM2 calculations | [70] |
| WT K103N Y181C | 3BGR 4I2Q 4I2P | PDB complexes | NNRTIs dihydro-pyrimidinone (DHPM) derivatives, probably NNRTIs | Molegro Virtual Docking | N/D | [71] |
| A set of various mutations | 1HNV | Computationally | NNRTI marine diterpenes | Autodock 4.2 | N/D | [72] |
| K103N | 3MED | PDB complex | NNRTI rilpivirine | AutodockRaccoon | Molecular dynamics using AMBER 12 (F99SB) force field | [73] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarasova, O.; Poroikov, V.; Veselovsky, A. Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review. Molecules 2018, 23, 1233. https://doi.org/10.3390/molecules23051233
Tarasova O, Poroikov V, Veselovsky A. Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review. Molecules. 2018; 23(5):1233. https://doi.org/10.3390/molecules23051233
Chicago/Turabian StyleTarasova, Olga, Vladimir Poroikov, and Alexander Veselovsky. 2018. "Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review" Molecules 23, no. 5: 1233. https://doi.org/10.3390/molecules23051233






