Evaluation of Transition Metal Complexes of Benzimidazole-Derived Scaffold as Promising Anticancer Chemotherapeutics
Abstract
:1. Introduction
2. Results and Discussion
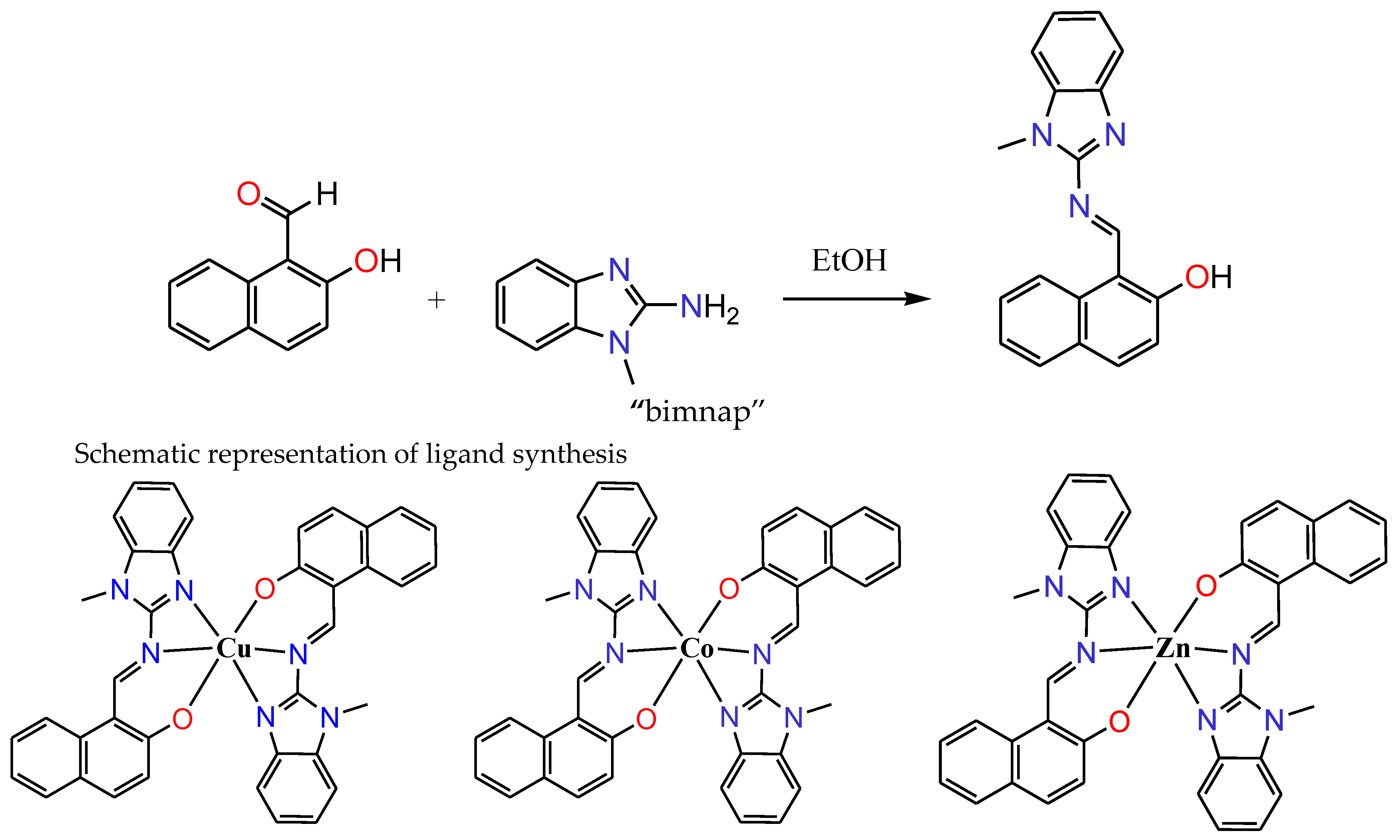
2.1. Synthesis and Characterization
2.2. X-ray Crystallography
2.3. DNA Binding Studies
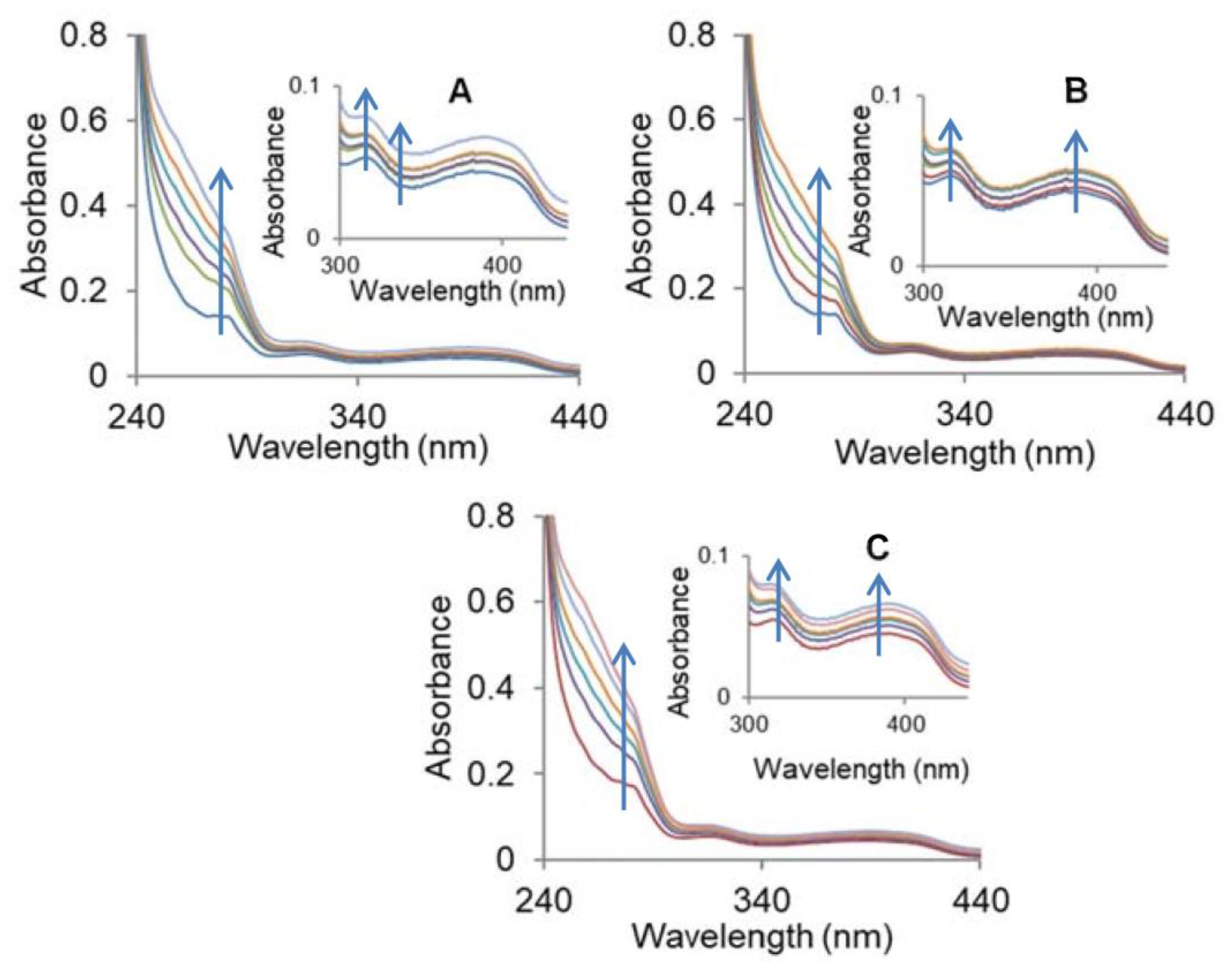
2.3.1. Absorption Studies
2.3.2. Fluorescence Quenching Experiments
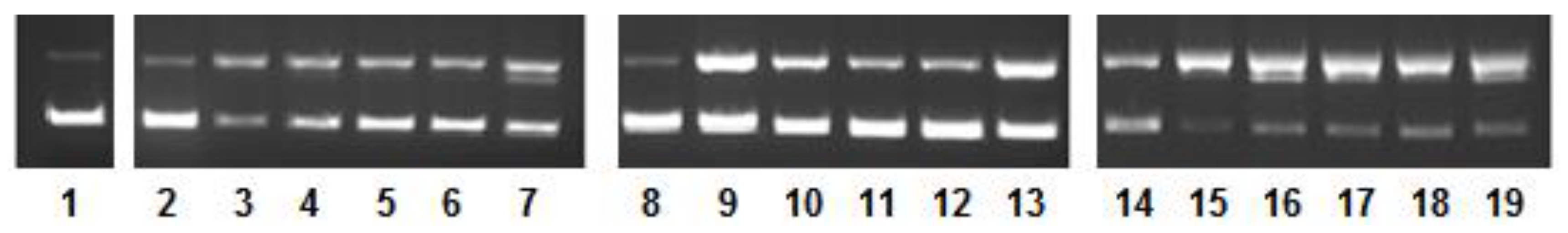
2.4. DNA Nuclease Activity and Mechanistic Pathways
2.5. In Vitro Cytotoxicity
2.5.1. Analysis of Growth Inhibition Using MTT Assay
2.5.2. Effect on Cancer Cell Adhesion and Cell Migration
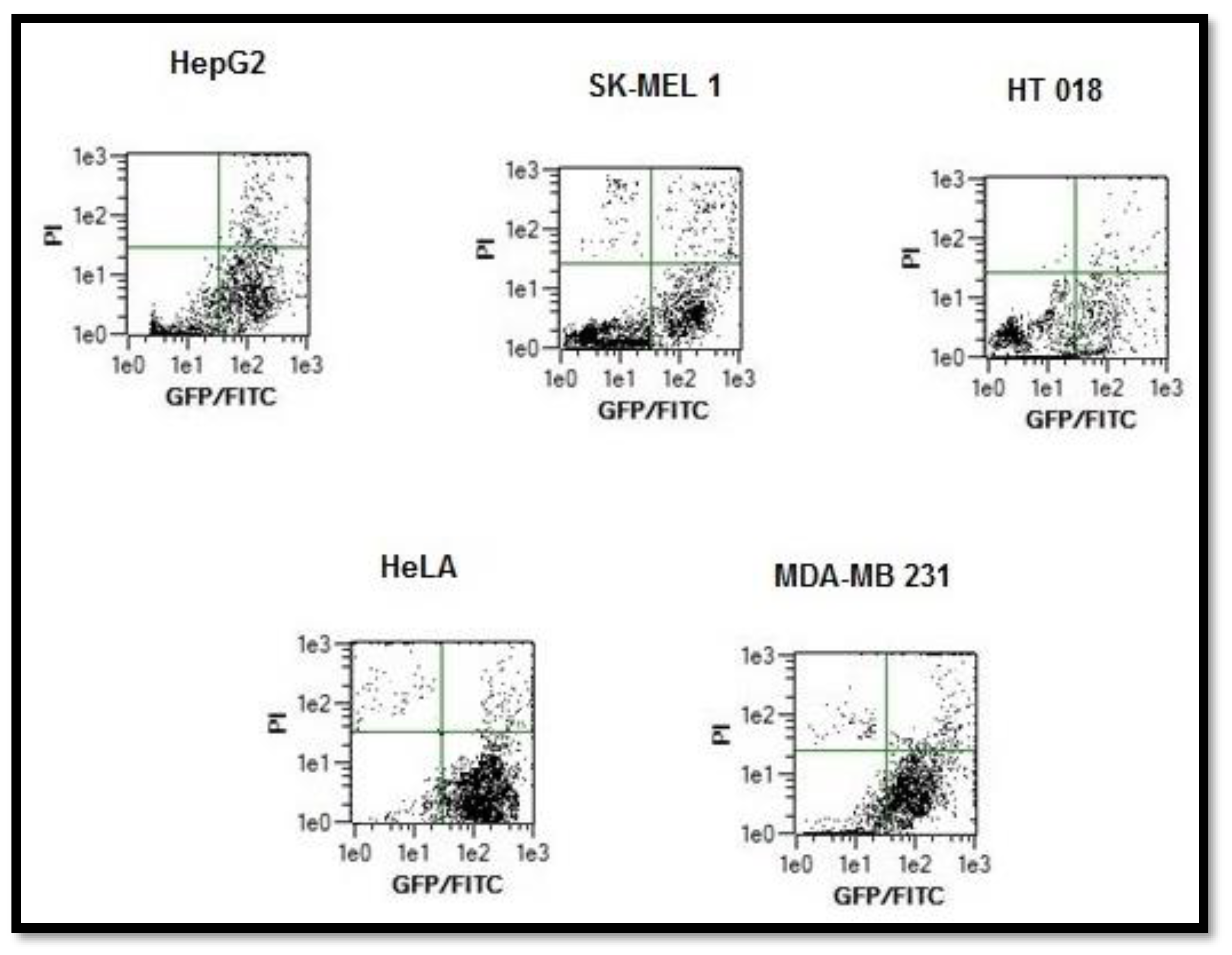
2.5.3. Annexin V Apoptosis Detection Assay
2.6. In Vivo Toxicity of Complexes 1–3
2.6.1. Hematological Study
2.6.2. Effect of Complexes 1–3 on Liver Function Tests
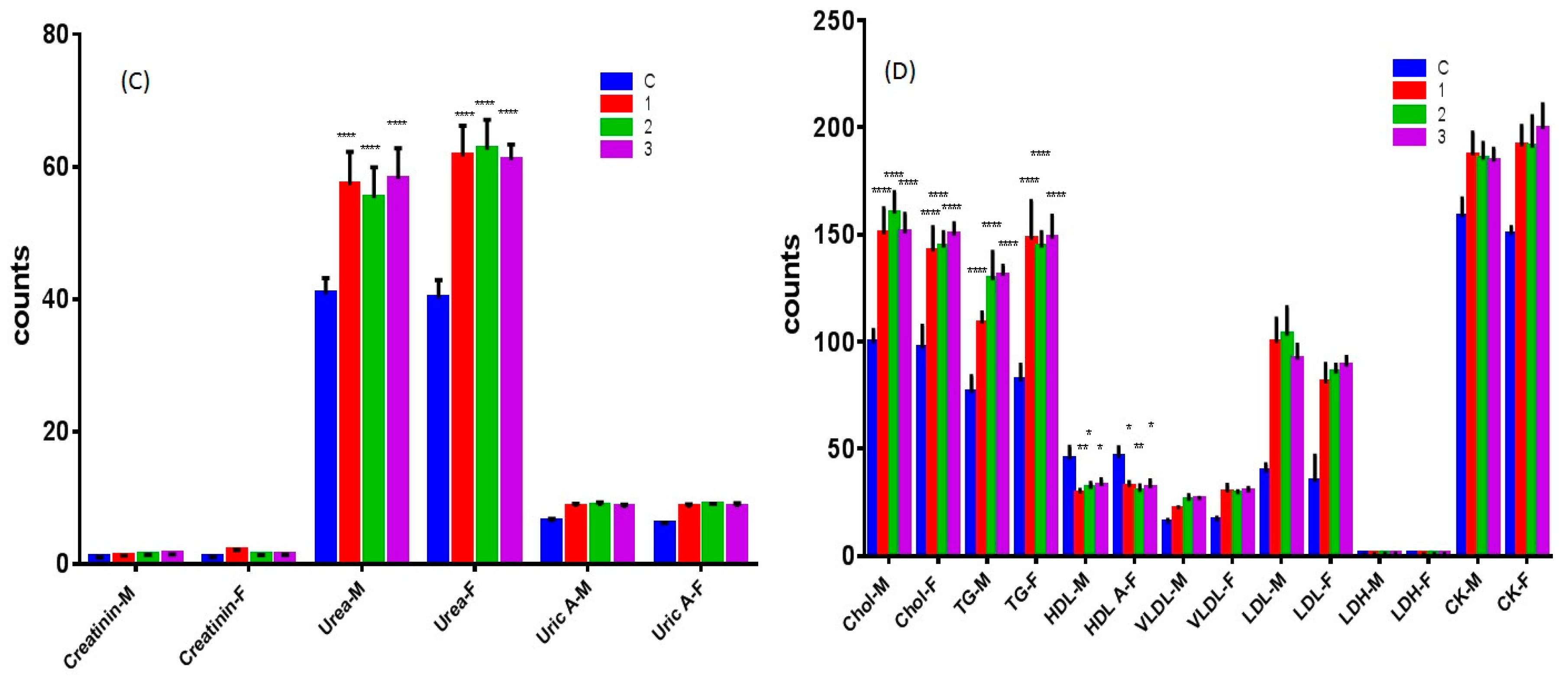
2.6.3. Nephrotoxicity Tests of Complexes 1–3
2.6.4. Effect of Complexes 1–3 on Lipid and Heart Function
3. Experimental Section
3.1. Materials and Methods
3.2. Data Collection
3.3. Synthesis of Ligand “Bimnap”
3.4. Synthetic Procedure
3.4.1. Copper Complex (1)
3.4.2. Cobalt Complex (2)
3.4.3. Zinc Complex (3)
3.5. DNA Binding and Nuclease Activity
3.6. Cell Lines and Culture Conditions
3.7. MTT Assay
3.8. Measurement of Cancer Cell Adhesion
3.9. Measurement of Cancer Cell Migration
3.10. Analysis of Annexin-V Binding by Flow Cytometry
3.11. In Vivo Toxicity
3.11.1. Toxicity Study Design
3.11.2. Chronic Toxicity Study
3.11.3. Hematological Studies
3.11.4. Serum Analysis of Biochemical Parameters
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wong, E.; Giandomenico, C.M. Current status of platinum-based antitumor drugs. Chem. Rev. 1999, 99, 2451–2466. [Google Scholar] [CrossRef] [PubMed]
- Alderden, R.A.; Hall, M.D.; Hambley, T.W. The discovery and development of cisplatin. J. Chem. Educ. 2006, 83, 728. [Google Scholar] [CrossRef]
- Jamieson, E.R.; Lippard, S.J. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, C.R.; Suntharalingam, K. Advances in cobalt complexes as anticancer agents. Dalton Trans. 2015, 44, 13796–13808. [Google Scholar] [CrossRef] [PubMed]
- Bergamo, A.; Sava, G. Ruthenium anticancer compounds: Myths and realities of the emerging metal-based drugs. Dalton Trans. 2011, 40, 7817–7823. [Google Scholar] [CrossRef] [PubMed]
- Bergamo, A.; Gaiddon, C.; Schellens, J.H.M.; Beijnen, J.H.; Sava, G. Approaching tumour therapy beyond platinum drugs: Status of the art and perspectives of ruthenium drug candidates. J. Inorg. Biochem. 2012, 106, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Romero-Canelón, I.; Sadler, P.J. Next-Generation Metal Anticancer Complexes: Multitargeting via Redox Modulation. Inorg. Chem. 2013, 52, 12276–12291. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, L.; Kawabata, T.; Yoshino, T.; Yang, B.B.; Okada, S. Cupric itrilotriacetate induces oxidative DNA damage and apoptosis in human leukemia HL-60 cells. Free Radic. Biol. Med. 1998, 25, 568–575. [Google Scholar] [CrossRef]
- Liang, F.; Wu, C.; Lin, H.; Li, T.; Gao, D.; Li, Z.; Wei, J.; Zheng, C.; Sun, M. Copper complex of hydroxyl-Substituted triazamacrocyclic ligand and its antitumor activity. Bioorg. Med. Chem. Lett. 2003, 13, 2469–2472. [Google Scholar] [CrossRef]
- Easmon, J.; Purstinger, G.; Heinisch, G.; Roth, T.; Fiebig, H.H.; Holzer, W.; Jager, W.; Jenny, M.; Hofmann, J. Synthesis, cytotoxicity, and antitumor activity of copper(II) and iron(II) complexes of 4N-azabicyclo[3.2.2]nonane thiosemicarbazones derived from acyl diazines. J. Med. Chem. 2001, 44, 2164–2171. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, S.; Amir, S.; Arjmand, F.; Pettinari, C.; Marchetti, F.; Masciocchi, N.; Lupidi, G.; Pettinari, R. Mixed-ligand Cu(II)–vanillin Schiff base complexes; effect of coligands on their DNA binding, DNA cleavage, SOD mimetic and anticancer activity. Eur. J. Med. Chem. 2013, 60, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Manikandamathavan, V.M.; Rajapandian, V.; Freddy, A.J.; Weyhermuller, T.; Subramanian, V.; Nair, B.U. Synthesis, DNA binding and antileishmanial activity of low molecular weight bis-arylimidamides. Eur. J. Med. Chem. 2012, 57, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Adman, E.T. Copper protein structures. Adv. Protein Chem. 1991, 42, 145–147. [Google Scholar] [PubMed]
- Choi, M.; Sukumar, N.; Liu, A.; Davidson, V.L. Defining the Role of the Axial Ligand of the Type 1 Copper Site in Amicyanin by Replacement of Methionine with Leucine. Biochemistry 2009, 48, 9174–9184. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Ghorai, P.; Brandao, P.; Ghosh, D.; Bhuiya, S.; Chattopadhyay, D.; Das, S.; Saha, A. Syntheses, crystal structures, DNA binding, DNA cleavage, molecular docking and DFT study of Cu(II) complexes involving N2O4 donor azo Schiff base ligands. New J. Chem. 2018, 42, 246–259. [Google Scholar] [CrossRef]
- Tapiero, H.; Tew, K.D. Trace elements in human physiology and pathology: Zinc and metallothioneins. Biomed. Pharmacother. 2003, 57, 399–411. [Google Scholar] [CrossRef]
- Walkup, G.K.; Burdette, S.C.; Lippard, S.J.; Tsien, R.Y. A New cell-permeable fluorescent probe for Zn2+. J. Am. Chem. Soc. 2000, 122, 5644–5645. [Google Scholar] [CrossRef]
- Vallee, B.L.; Auld, D.S. New perspective on zinc biochemistry: Cocatalytic sites in multi-zinc enzymes. Biochemistry 1993, 32, 6493–6500. [Google Scholar] [CrossRef] [PubMed]
- Fusch, E.C.; Lippert, B. [Zn3(OH)2(1-MeC-N3)5(1-MeC-O2)3]4+ (1-MeC =1-Methylcytosine): Structural model for DNA crosslinking and DNA rewinding by Zn(II)? J. Am. Chem. Soc. 1994, 116, 7204–7209. [Google Scholar] [CrossRef]
- Sharif, R.; Thomas, P.; Zalewski, P.; Fenech, M. Zinc deficiency or excess within the physiological range increases genome instability and cytotoxicity respectively, in human oral keratinocyte cells. Genes Nutr. 2012, 7, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, D.K.; Chadha, V.D. Zinc: A promising agent in dietary chemoprevention of cancer. Indian J. Med. Res. 2010, 132, 676–682. [Google Scholar] [PubMed]
- Liu, S.; Cao, W.; Yu, L.; Zheng, W.; Li, L.; Fan, C.; Chen, T. Zinc(II) complexes containing bis-benzimidazole derivatives as a new class of apoptosis inducers that trigger DNA damage-mediated p53 phosphorylation in cancer cells. Dalton Trans. 2013, 42, 5932–5940. [Google Scholar] [CrossRef] [PubMed]
- Ott, I.; Gust, R. Non platinum metal complexes as anti-cancer drugs. Arch. Pharm. 2007, 340, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Banerjee, K.; Sinha, A.; Das, S.; Majumder, S.; Majumdar, S.; Choudhuri, S.K. A zinc Schiff base complex inhibits cancer progression both in vivo and in vitro by inducing apoptosis. Environ. Toxicol. Pharmacol. 2017, 56, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Renfrew, A.K.; Neill, E.S.; Hambley, T.W.; New, E.J. Harnessing the properties of cobalt coordination complexes for biological application. Coord. Chem. Rev. 2017. [Google Scholar] [CrossRef]
- Delaney, S.; Pascaly, M.; Bhattacharya, P.K.; Han, K.; Barton, J.K. Oxidative damage by ruthenium complexes containing the dipyridophenazine ligand or Its erivatives: A focus on intercalation. Inorg. Chem. 2002, 41, 1966–1974. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Zaki, M.; Khan, R.A.; Alsalme, A.; Ahmad, M.; Tabassum, S. Coumarin centered copper(II) complex with appended-imidazole as cancer chemotherapeutic agents against lung cancer: Molecular insight via DFT-based vibrational analysis. RSC Adv. 2017, 7, 36056–36071. [Google Scholar] [CrossRef]
- Khan, R.A.; Yadav, S.; Hussain, Z.; Arjmand, F.; Tabassum, S. Carbohydrate linked organotin(IV) complexes as human topoisomerase Iα inhibitor and their antiproliferative effects against the human carcinoma cell line. Dalton Trans. 2014, 43, 2534–2548. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, F.; Aziz, M. Synthesis and characterization of dinuclear macrocyclic cobalt(II), copper(II) and zinc(II) complexes derived from 2,2,2′,2′-S,S[bis(bis-N,N-2-thiobenzimidazolyloxalato-1,2-ethane)]: DNA binding and cleavage studies. Eur. J. Med. Chem. 2009, 44, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Uma, V.; Elango, M.; Nair, B.U. Copper(II) Terpyridine Complexes: Effect of Substituent on DNA Binding and Nuclease Activity. Eur. J. Inorg. Chem. 2007, 2007, 3484–3490. [Google Scholar] [CrossRef]
- Sharma, S.; Toupet, L.; Arjmand, F. De novo design of a hydrolytic DNA cleavage agent, mono nitratobis(phen)cobalt(II) aqua nitrate complex. New J. Chem. 2017, 41, 2883–2886. [Google Scholar] [CrossRef]
- Parveen, S.; Arjmand, F.; Mohapatra, D.K. Zinc(II) complexes of Pro-Gly and Pro-Leu dipeptides: Synthesis, characterization, in vitro DNA binding and cleavage studies. J. Photochem. Photobiol. B 2013, 126, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Jun, T.; Bochu, W.; Zhu, L. Hydrolytic cleavage of DNA by quercetin zinc(II) complex. Bioorg. Med. Chem. Lett. 2007, 17, 1197–1199. [Google Scholar] [CrossRef] [PubMed]
- Massoud, S.S.; Perkins, R.S.; Louka, F.R.; Xu, W.; Le Roux, A.; Dutercq, Q.; Fischer, R.C.; Mautner, F.A.; Handa, M.; Hiraoka, Y.; et al. Efficient hydrolytic cleavage of plasmid DNA by chloro-cobalt(II) complexes based on sterically hindered pyridyl tripod tetraamine ligands: Synthesis, crystal structure and DNA cleavage. Dalton Trans. 2014, 43, 10086–10103. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Hail, N., Jr.; Lotan, R. Apoptosis as a novel target for cancer chemoprevention. J. Natl. Cancer Inst. 2004, 96, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Van Zutphen, S.; Reedijk, J. Targeting platinum anti-tumour drugs: Overview of strategies employed to reduce systemic toxicity. Coord. Chem. Rev. 2005, 249, 2845–2853. [Google Scholar] [CrossRef]
- Boulikas, T.; Pantos, A.; Bellis, E.; Christofis, P. Designing platinum compounds in cancer: Structures and mechanisms. Cancer Ther. 2007, 5, 537–583. [Google Scholar]
- Tabassum, S.; Asim, A.; Khan, R.A.; Arjmand, F.; Divya, R.; Balaji, P.; Akbarsha, M.A. A multifunctional molecular entity CuII-SnIV heterobimetallic complex as potential cancer chemotherapeutic agents: DNA binding/cleavage, SOD mimetic, topoisomerase Iα inhibitory and in vitro cytotoxic activities. RSC Adv. 2015, 5, 47439–47450. [Google Scholar] [CrossRef]
- Usman, M.; Arjmand, F.; Khan, R.A.; Alsalme, A.; Ahmad, M.; Bishwas, M.S.; Tabassum, S. Tetranuclear Cubane Cu4O4 complexes as prospective Anticancer Agents: Design, Synthesis, Structural Elucidation, Magnetism, Computational Studies and Cytotoxicity Assays. Inorg. Chim. Acta 2018, 473, 121–132. [Google Scholar] [CrossRef]
- Khan, A.A.; Jabeen, M.; Chauhan, A.; Owais, M. Synthesis, and characterization of novel n-9 fatty acid conjugates possessing antineoplastic properties. Lipids 2012, 47, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Alanazi, A.M.; Jabeen, M.; Chauhan, A.; Abdelhameed, A.S. Design, Synthesis and In Vitro Anticancer Evaluation of a Stearic Acid-based Ester Conjugate. Anticancer Res. 2013, 33, 1217–1224. [Google Scholar]
- Khan, A. Pro-apoptotic activity of nano-escheriosome based oleic acid conjugate against 7,12-dimethylbenz(a)anthracene (DMBA) induced cutaneous carcinogenesis. Biomed. Pharmacother. 2017, 90, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.H.; Qureshi, S.; Tariq, M.; Ageel, A.M. Toxicity studies on six plants used in the traditional Arab system of medicine. Phytother. Res. 1989, 3, 25–29. [Google Scholar] [CrossRef]
- World Health Organization (WHO) Scientific Group. Principles for Pre-Clinical Testing of Drugs Safety; Technical Report Series; World Health Organization: Geneva, Switzerland, 1967; Volume 341, pp. 9–11. [Google Scholar]
- Edwards, C.R.W.; Bouchier, I.A.D. Davidson’s Principles and Practice Medicine; Churchill Livingstone Press: London, UK, 1991; p. 492. [Google Scholar]
- Daniel, W.W. Biostatisitics: A Foundation for Analysis in the Health Sciences, 6th ed.; Wiley: New York, NY, USA, 1995; pp. 273–303. [Google Scholar]
Sample Availability: Samples of the compounds will be available on request from the authors. |
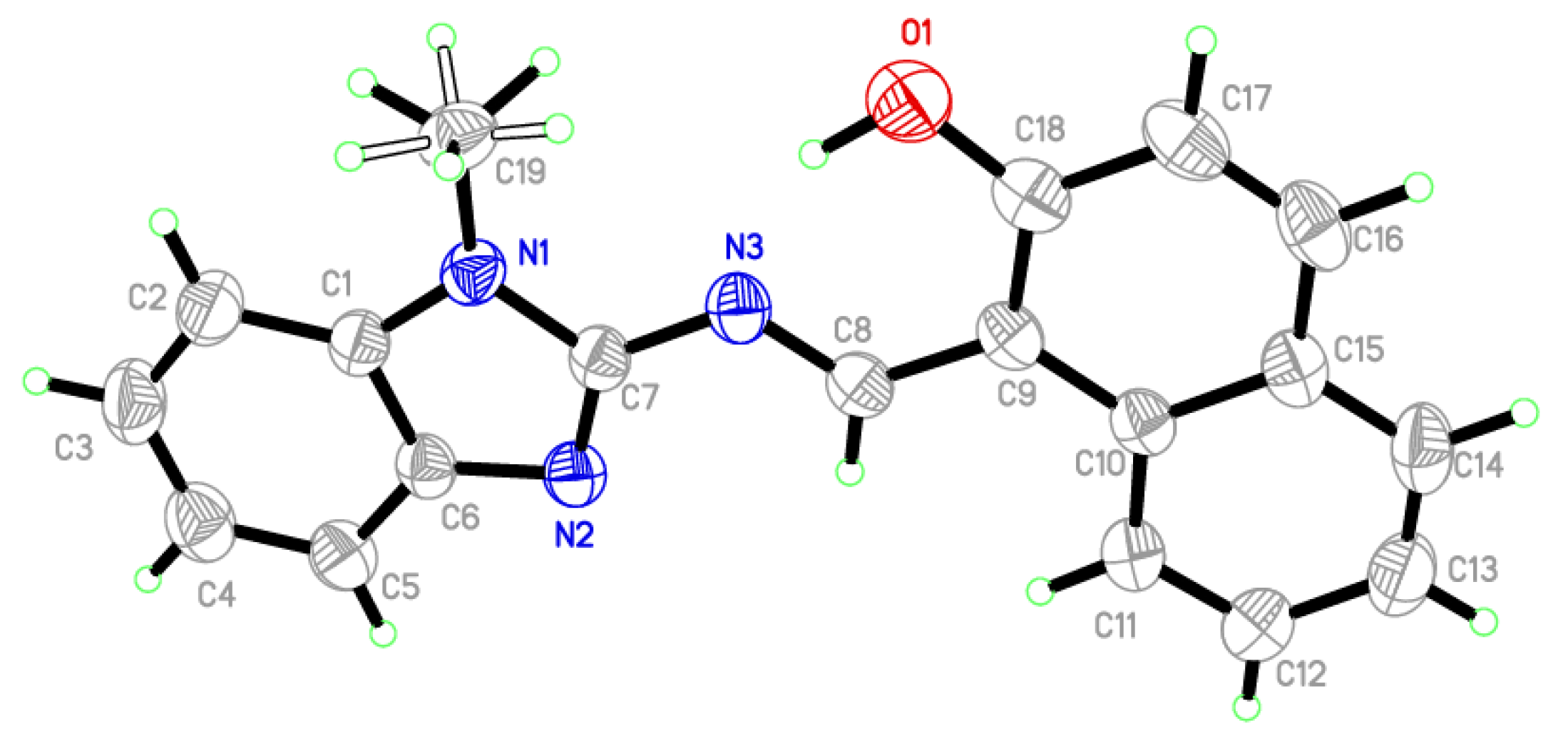



| Parameters | Ligand (bimnap) |
|---|---|
| Formula | C19H15N3O |
| Fw (g mol–1) | 301.34 |
| crystal system | Monoclinic |
| space group | P21/c |
| a (Å) | 9.4568(6) |
| b (Å) | 12.9529(10) |
| c (Å) | 13.7156(8) |
| β (deg) | 116.171(4) |
| U (Å3) | 1507.83(18) |
| Z | 4 |
| µ (mm–1) | 0.085 |
| D (g cm−3) | 1.327 |
| F(000) | 632 |
| Temp (K) | 293(2) |
| measured reflns | 21459 |
| unique reflns | 4181 |
| Ɵ range of data (°) | 2.28–21.53 |
| Goodness of fit on F2 | 1.035 |
| Final Rb indices | R1 = 0.053 |
| [I > 2σ(I)] | wR2 = 0.122 |
| CCDC | 1485309 |
| Bond Angle (°) | Bond Length (Å) | |||
|---|---|---|---|---|
| C18-O1-H1O1 | 108.1 (18) | N3-C8-C9-C18 | −1.5 (3) | |
| N3-C8-C9-C10 | 179.09 (19) | C8-C9-C18-O1 | −1.5 (3) | |
| C19-N1-C7-N2 | 173.3 (2) | C19-N1-C1-C2 | 7.3 (4) | |
| C1-N1-C7-N3 | 177.67 (17) | O1-C18 | 1.341 (3) | |
| C8-N3-C7-N1 | −178.96 (17) | O1-H1O1 | 0.97 (3) | |
| N1-C19-H19B | 112 (2) | N1-C7 | 1.368 (2) | |
| N1-C19-H19A | 107 (3) | N1-C1 | 1.380 (3) | |
| O1-C18-C9 | 122.3 (2) | N1-C19 | 1.453 (3) | |
| O1-C18-C17 | 116.6 (2) | N2-C7 | 1.316 (3) | |
| N2-C7-N1 | 114.16 (19) | N2-C6 | 1.385 (3) | |
| N2-C7-N3 | 128.24 (19) | N3-C8 | 1.296 (2) | |
| N3-C8-C9 | 121.3 (2) | N1-C19-H19C | 108 (2) | |
| Sample Code | HepG2 | SK-MEL-1 | HT018 | HeLa | MDA-MB 231 |
|---|---|---|---|---|---|
| (Liver) (µM) | (Skin) (µM) | (Colon) (µM) | (Cervical) (µM) | (Breast) (µM) | |
| Complex 1 | 26 ± 2.2 (Less potent) | 54.7 ± 3.2 (Less potent) | 29 ± 3.1 (Less potent) | 23 ± 2.2 | 22 ± 2.4 |
| Complex 2 | 45 ± 2.6 (Less potent) | 38 ± 2.3 (Less potent) | 51.3 ± 2.3 (Less potent) | 44.5 ± 2.2 (Less potent) | 54.6 ± 2.6 (Less potent) |
| Complex 3 | 12 ± 3.1 | 11.1 ± 2.3 | 15.8 ± 1.9 | 8.3 ± 1.5 | 6.66 ± 1.7 |
| Ligand | NA | NA | NA | NA | NA |
| Vehicle control (0.1% DMSO) | NA | NA | NA | NA | NA |
| Cisplatin | 6 ± 0.4 | 5.6 ± 0.8 | 5.7 ± 0.2 | 6 ± 0.6 | 3.1 ± 0.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, A.; AlAjmi, M.F.; Rehman, M.T.; Khan, A.A.; Shaikh, P.A.; Khan, R.A. Evaluation of Transition Metal Complexes of Benzimidazole-Derived Scaffold as Promising Anticancer Chemotherapeutics. Molecules 2018, 23, 1232. https://doi.org/10.3390/molecules23051232
Hussain A, AlAjmi MF, Rehman MT, Khan AA, Shaikh PA, Khan RA. Evaluation of Transition Metal Complexes of Benzimidazole-Derived Scaffold as Promising Anticancer Chemotherapeutics. Molecules. 2018; 23(5):1232. https://doi.org/10.3390/molecules23051232
Chicago/Turabian StyleHussain, Afzal, Mohamed F. AlAjmi, Md. Tabish Rehman, Azmat Ali Khan, Perwez Alam Shaikh, and Rais Ahmad Khan. 2018. "Evaluation of Transition Metal Complexes of Benzimidazole-Derived Scaffold as Promising Anticancer Chemotherapeutics" Molecules 23, no. 5: 1232. https://doi.org/10.3390/molecules23051232







