Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein
Abstract
:1. Introduction
2. Results
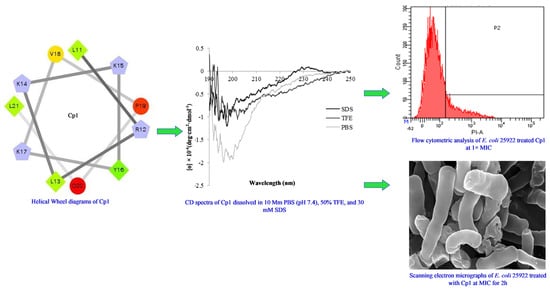
2.1. Characterization and Structure Variability of the Peptides
2.2. Activities of the Peptides
2.2.1. Antimicrobial Activity
2.2.2. Hemolytic Activity
2.2.3. Cytotoxicity Activity
2.2.4. Salt Sensitivity
2.3. Mechanism of Action of the Peptides
2.3.1. Evaluation of Outer Membrane Permeability
2.3.2. Evaluation of Inner Membrane Permeability
2.3.3. Cytoplasmic Membrane Electrical Potential
2.3.4. Flow Cytometry
2.3.5. SEM
2.3.6. TEM
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Assay
4.2. Hemolytic Activity
4.3. Cytotoxicity
4.4. Salt Sensitivity Assays
4.5. Circular Dichroism (CD) Measurements
4.6. Outer Membrane Permeability Assay
4.7. Inner Membrane Permeability Assay
4.8. Cytoplasmic Membrane Depolarization Assay
4.9. Flow Cytometry
4.10. Scanning Electron Microscopy (SEM)
4.11. Transmission Electron Microscopy (TEM)
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Soblosky, L.; Ramamoorthy, A.; Chen, Z. Membrane interaction of antimicrobial peptides using E. coli lipid extract as model bacterial cell membranes and SFG spectroscopy. Chem. Phys. Lipids 2015, 187, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-L.; Shi, Y.-H.; Zhao, W.; Hao, G.; Le, G.-W. Discovery of a novel antimicrobial peptide using membrane binding-based approach. Food Control 2009, 20, 149–156. [Google Scholar] [CrossRef]
- Perez Espitia, P.J.; de Fátima Ferreira Soares, N.; dos Reis Coimbra, J.S.; de Andrade, N.J.; Souza Cruz, R.; Medeiros, A.; Antonio, E. Bioactive peptides: Synthesis, properties, and applications in the packaging and preservation of food. Compr. Rev. Food Sci. Food Saf. 2012, 11, 187–204. [Google Scholar] [CrossRef]
- Wu, M.; Hancock, R.E. Improved derivatives of bactenecin, a cyclic dodecameric antimicrobial cationic peptide. Antimicrob. Agents Chemother. 1999, 43, 1274–1276. [Google Scholar] [PubMed]
- Dong, N.; Ma, Q.; Shan, A.; Cao, Y. Design and biological activity of β-hairpin-like antimicrobial peptide. Chin. J. Biotechnol. 2012, 28, 243–250. [Google Scholar]
- Dennison, S.R.; Mura, M.; Harris, F.; Morton, L.H.; Zvelindovsky, A.; Phoenix, D.A. The role of C-terminal amidation in the membrane interactions of the anionic antimicrobial peptide, maximin H5. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Natsuga, K.; Cipolat, S.; Watt, F.M. Increased bacterial load and expression of antimicrobial peptides in skin of barrier-deficient mice with reduced cancer susceptibility. J. Investig. Dermatol. 2016, 136, 99–106. [Google Scholar] [CrossRef] [PubMed]
- De Souza Cândido, E.; Sousa, D.A.; Viana, J.C.; de Oliveira-Júnior, N.G.; Miranda, V.; Franco, O.L. The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides 2014, 55, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Nogués, V.M.; Boix, E. A theoretical approach to spot active regions in antimicrobial proteins. BMC Bioinform. 2009, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Barzyk, W.; Campagna, S.; Więcław, K.; Korchowiec, B.; Rogalska, E. The affinity of two antimicrobial peptides derived from bovine milk proteins for model lipid membranes. Colloids Surf. A Physicochem. Eng. Asp. 2009, 343, 104–110. [Google Scholar] [CrossRef]
- Dong, N.; Ma, Q.; Shan, A.; Lv, Y.; Hu, W.; Gu, Y.; Li, Y. Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine-and valine-rich β-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother. 2012, 56, 2994–3003. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Hwang, J.-S.; Lee, J.; Kim, J.I.; Lee, D.G. Scolopendin 2, a cationic antimicrobial peptide from centipede, and its membrane-active mechanism. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Lv, Y.; Gu, Y.; Dong, N.; Li, D.; Shan, A. Rational design of cationic antimicrobial peptides by the tandem of leucine-rich repeat. Amino Acids 2013, 44, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shiell, B.; Wan, J.; Coventry, M.; Michalski, W.; Lee, A.; Roginski, H. The molecular characterisation and antimicrobial properties of amidated bovine β-lactoglobulin. Int. Dairy J. 2007, 17, 1450–1459. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. Fungicidal mechanisms of the antimicrobial peptide Bac8c. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 673–679. [Google Scholar] [CrossRef] [PubMed]
- McClean, S.; Beggs, L.B.; Welch, R.W. Antimicrobial activity of antihypertensive food-derived peptides and selected alanine analogues. Food Chem. 2014, 146, 443–447. [Google Scholar] [CrossRef] [PubMed]
- López-Expósito, I.; Gómez-Ruiz, J.Á.; Amigo, L.; Recio, I. Identification of antibacterial peptides from ovine α s2-casein. Int. Dairy J. 2006, 16, 1072–1080. [Google Scholar] [CrossRef]
- López-Expósito, I.; Amigo, L.; Recio, I. Identification of the initial binding sites of α s2-casein f (183–207) and effect on bacterial membranes and cell morphology. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 2444–2449. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.; Silva, F.; Egito, A.; Silva, T.; Lima-Filho, J.; Porto, A.; Moreira, K. New peptides obtained by hydrolysis of caseins from bovine milk by protease extracted from the latex Jacaratia corumbensis. LWT Food Sci. Technol. 2012, 49, 73–79. [Google Scholar] [CrossRef]
- McCann, K.; Shiell, B.; Michalski, W.; Lee, A.; Wan, J.; Roginski, H.; Coventry, M. Isolation and characterisation of a novel antibacterial peptide from bovine α S1-casein. Int. Dairy J. 2006, 16, 316–323. [Google Scholar] [CrossRef]
- Tang, W.; Yuan, H.; Zhang, H.; Wang, L.; Qian, H.; Qi, X. An antimicrobial peptide screened from casein hydrolyzate by Saccharomyces cerevisiae cell membrane affinity method. Food Control 2015, 50, 413–422. [Google Scholar] [CrossRef]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef]
- Yan, H.; Li, S.; Sun, X.; Mi, H.; He, B. Individual substitution analogs of Mel (12–26), melittin’s C-terminal 15-residue peptide: Their antimicrobial and hemolytic actions. FEBS Lett. 2003, 554, 100–104. [Google Scholar] [CrossRef]
- Pandey, B.K.; Ahmad, A.; Asthana, N.; Azmi, S.; Srivastava, R.M.; Srivastava, S.; Verma, R.; Vishwakarma, A.L.; Ghosh, J.K. Cell-selective lysis by novel analogues of melittin against human red blood cells and Escherichia coli. Biochemistry 2010, 49, 7920–7929. [Google Scholar] [CrossRef] [PubMed]
- Asthana, N.; Yadav, S.P.; Ghosh, J.K. Dissection of antibacterial and toxic activity of Melittin a leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J. Biol. Chem. 2004, 279, 55042–55050. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Shan, A.S.; Dong, N.; Gu, Y.; Sun, W.Y.; Hu, W.N.; Feng, X.J. Cell selectivity and interaction with model membranes of Val/Arg-rich peptides. J. Pept. Sci. 2011, 17, 520. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tan, T.; Xu, W.; Xu, L.; Dong, N.; Ma, D.; Shan, A. Rational design of mirror-like peptides with alanine regulation. Amino Acids 2016, 48, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Oren, Z.; Shai, Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: Structure-function study. Biochemistry 1997, 36, 1826–1835. [Google Scholar] [CrossRef] [PubMed]
- González, R.; Gentina, J.C.; Acevedo, F. Optimisation of the solids suspension conditions in a continuous stirred tank reactor for the biooxidation of refractory gold concentrates. Electron. J. Biotechnol. 2003, 6, 233–243. [Google Scholar]
- Hancock, R.E.; Brown, K.L.; Mookherjee, N. Host defence peptides from invertebrates—Emerging antimicrobial strategies. Immunobiology 2006, 211, 315–322. [Google Scholar] [CrossRef] [PubMed]
- LaRock, C.N.; Nizet, V. Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 3047–3054. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Rozek, A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 2002, 206, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Shao, C.; Wang, J.; Shan, A.; Xu, L.; Dong, N.; Li, Z. Short, multiple-stranded β-hairpin peptides have antimicrobial potency with high selectivity and salt resistance. Acta Biomater. 2016, 30, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.J.; Schibli, D.J.; Jing, W.; Lohmeier-Vogel, E.M.; Epand, R.F.; Epand, R.M. Towards a structure-function analysis of bovine lactoferricin and related tryptophan-and arginine-containing peptides. Biochem. Cell Biol. 2002, 80, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chou, S.; Wang, J.; Shao, C.; Li, W.; Zhu, X.; Shan, A. Antimicrobial activity and membrane-active mechanism of tryptophan zipper-like β-hairpin antimicrobial peptides. Amino Acids 2015, 47, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, L.; Wang, J.; Ma, Z.; Xu, W.; Li, J.; Shan, A. Characterization of antimicrobial activity and mechanisms of low amphipathic peptides with different α-helical propensity. Acta Biomater. 2015, 18, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.; Ross, R.; Fitzgerald, G.; Hill, C.; Stanton, C. Casein-derived antimicrobial peptides generated by Lactobacillus acidophilus DPC6026. Appl. Environ. Microbiol. 2006, 72, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Ding, J.; Li, H.; Li, L.; Zhao, R.; Shen, Z.; Fan, X.; Xi, T. Effects of cations and pH on antimicrobial activity of thanatin and s-thanatin against Escherichia coli ATCC25922 and B. subtilis ATCC 21332. Curr. Microbiol. 2008, 57, 552–557. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, M.J.; Rivas, L.; Burgess, C.M.; Fanning, S.; Duffy, G. Inhibition of verocytotoxigenic Escherichia coli by antimicrobial peptides caseicin A and B and the factors affecting their antimicrobial activities. Int. J. Food Microbiol. 2012, 153, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, W.; Takase, M.; Wakabayashi, H.; Kawase, K.; Tomita, M. Antibacterial spectrum of lactoferricin B, a potent bactericidal peptide derived from the N-terminal region of bovine lactoferrin. J. Appl. Bacteriol. 1992, 73, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.L.; Sun, Y.; Liu, H.; Lehrer, R.I.; Shafer, W.M. Susceptibility of Treponema pallidum to host-derived antimicrobial peptides. Peptides 2003, 24, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.M.; Darouiche, R.O.; Legarda, D.; Connell, N.; Diamond, G. Characterization of a fish antimicrobial peptide: Gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 2000, 44, 2039–2045. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Zhu, X.; Chou, S.; Shan, A.; Li, W.; Jiang, J. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 2014, 35, 8028–8039. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hao, D.; Chen, Y.; Xu, Y.; Tan, J.; Huang, Y.; Li, F.; Chen, Y. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an α-helical antibacterial peptide against bacteria. Peptides 2011, 32, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Aquila, M.; Benedusi, M.; Koch, K.-W.; Dell’Orco, D.; Rispoli, G. Divalent cations modulate membrane binding and pore formation of a potent antibiotic peptide analog of alamethicin. Cell Calcium 2013, 53, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhu, X.; Tan, T.; Li, W.; Shan, A. Design of embedded-hybrid antimicrobial peptides with enhanced cell selectivity and anti-biofilm activity. PLoS ONE 2014, 9, e98935. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E. Bacterial membrane lipids: Where do we stand? Ann. Rev. Microbiol. 2003, 57, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Flores-Villaseñor, H.; Canizalez-Román, A.; Reyes-Lopez, M.; Nazmi, K.; de la Garza, M.; Zazueta-Beltrán, J.; León-Sicairos, N.; Bolscher, J.G. Bactericidal effect of bovine lactoferrin, LFcin, LFampin and LFchimera on antibiotic-resistant Staphylococcus aureus and Escherichia coli. Biometals 2010, 23, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, D.A.; Hurst, M.A.; Fujii, C.A.; Kung, A.; Ho, J.; Cheng, F.; Loury, D.J.; Fiddes, J.C. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 1997, 41, 1738–1742. [Google Scholar] [PubMed]
- Libardo, M.D.J.; Nagella, S.; Lugo, A.; Pierce, S.; Angeles-Boza, A.M. Copper-binding tripeptide motif increases potency of the antimicrobial peptide Anoplin via Reactive Oxygen Species generation. Biochem. Biophys. Res. Commun. 2015, 456, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.; Liu, L.-P.; Deber, C.M. Cationic hydrophobic peptides with antimicrobial activity. Antimicrob. Agents Chemother. 2002, 46, 3585–3590. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Mano, J.; Queiroz, J.; Gouveia, I. Incorporation of antimicrobial peptides on functionalized cotton gauzes for medical applications. Carbohydr. Polym. 2015, 127, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Schmidtchen, A.; Ringstad, L.; Kasetty, G.; Mizuno, H.; Rutland, M.W.; Malmsten, M. Membrane selectivity by W-tagging of antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Subbalakshmi, C.; Krishnakumari, V.; Nagaraj, R.; Sitaram, N. Requirements for antibacterial and hemolytic activities in the bovine neutrophil derived 13-residue peptide indolicidin. FEBS Lett. 1996, 395, 48–52. [Google Scholar] [CrossRef]
- Jacob, B.; Kim, Y.; Hyun, J.-K.; Park, I.-S.; Bang, J.-K.; Shin, S.Y. Bacterial killing mechanism of sheep myeloid antimicrobial peptide-18 (SMAP-18) and its Trp-substituted analog with improved cell selectivity and reduced mammalian cell toxicity. Amino Acids 2014, 46, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.L.; Rozek, A.; Patrzykat, A.; Hancock, R.E. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 2001, 276, 24015–24022. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kim, H.J.; Hahm, K.-S. Antibacterial synergism of novel antibiotic peptides with chloramphenicol. Biochem. Biophys. Res. Commun. 2004, 321, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Juba, M.L.; Porter, D.K.; Williams, E.H.; Rodriguez, C.A.; Barksdale, S.M.; Bishop, B.M. Helical cationic antimicrobial peptide length and its impact on membrane disruption. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, W.; Fan, M.; Tong, Z.; Kuang, R.; Jiang, W.; Ni, L. Antimicrobial and anti-biofilm effect of Bac8c on major bacteria associated with dental caries and Streptococcus mutans biofilms. Peptides 2014, 52, 61–67. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |









| Peptides | Sequence | Theoretical Mw (Da) | Measured Mw (Da) | Net Charge | H | μH | pI |
|---|---|---|---|---|---|---|---|
| Cp1 | LRLKKYKVPQL-NH2 | 1385.76 | 1384.79 | +4 | 0.345 | 0.096 | 10.46 |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-NH2 | 2847.49 | 2847.51 | +5 | 0.511 | 0.394 | 12.02 |
| Peptides | Minimum Inhibitory Concentration (MIC, μM) * | |
|---|---|---|
| Cp1 | Melittin | |
| Gram-negative bacteria | ||
| E. coli ATCC 25922 | 64 | 1 |
| E. coli UB1005 | 128 | 2 |
| Salmonella pullorum C7913 | 256 | 8 |
| Salmonella enterica subsp enterica CMCC 50071 | 256 | 2 |
| Gram-positive bacteria | ||
| Staphylococcus aureus ATCC 29213 | 640 | 2 |
| L. monocytogenes CMCC 54004 | 64 | 1 |
| MHC † | 32 | 1 |
| Peptides | Control | NaCl * | KCl * | NH4Cl * | MgCl2 * | CaCl2 * | FeCl3 * |
|---|---|---|---|---|---|---|---|
| Gram-negative strain E. coli 25922 | |||||||
| Cp1 | 64 | 64 | 64 | 64 | 128 | 128 | 64 |
| Melittin | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Gram-positive strain L. monocytogenes 54004 | |||||||
| Cp1 | 64 | 64 | 64 | 128 | 64 | 256 | 64 |
| Melittin | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Liu, Z.; Cao, S.; Wang, H.; Jiang, C.; Hussain, M.A.; Pang, S. Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein. Molecules 2018, 23, 1220. https://doi.org/10.3390/molecules23051220
Hou J, Liu Z, Cao S, Wang H, Jiang C, Hussain MA, Pang S. Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein. Molecules. 2018; 23(5):1220. https://doi.org/10.3390/molecules23051220
Chicago/Turabian StyleHou, Juncai, Zhijing Liu, Songsong Cao, Haimei Wang, Chenggang Jiang, Muhammad Altaf Hussain, and Shiyue Pang. 2018. "Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein" Molecules 23, no. 5: 1220. https://doi.org/10.3390/molecules23051220