The Late Embryogenesis Abundant Protein Family in Cassava (Manihot esculenta Crantz): Genome-Wide Characterization and Expression during Abiotic Stress
Abstract
:1. Introduction
2. Results
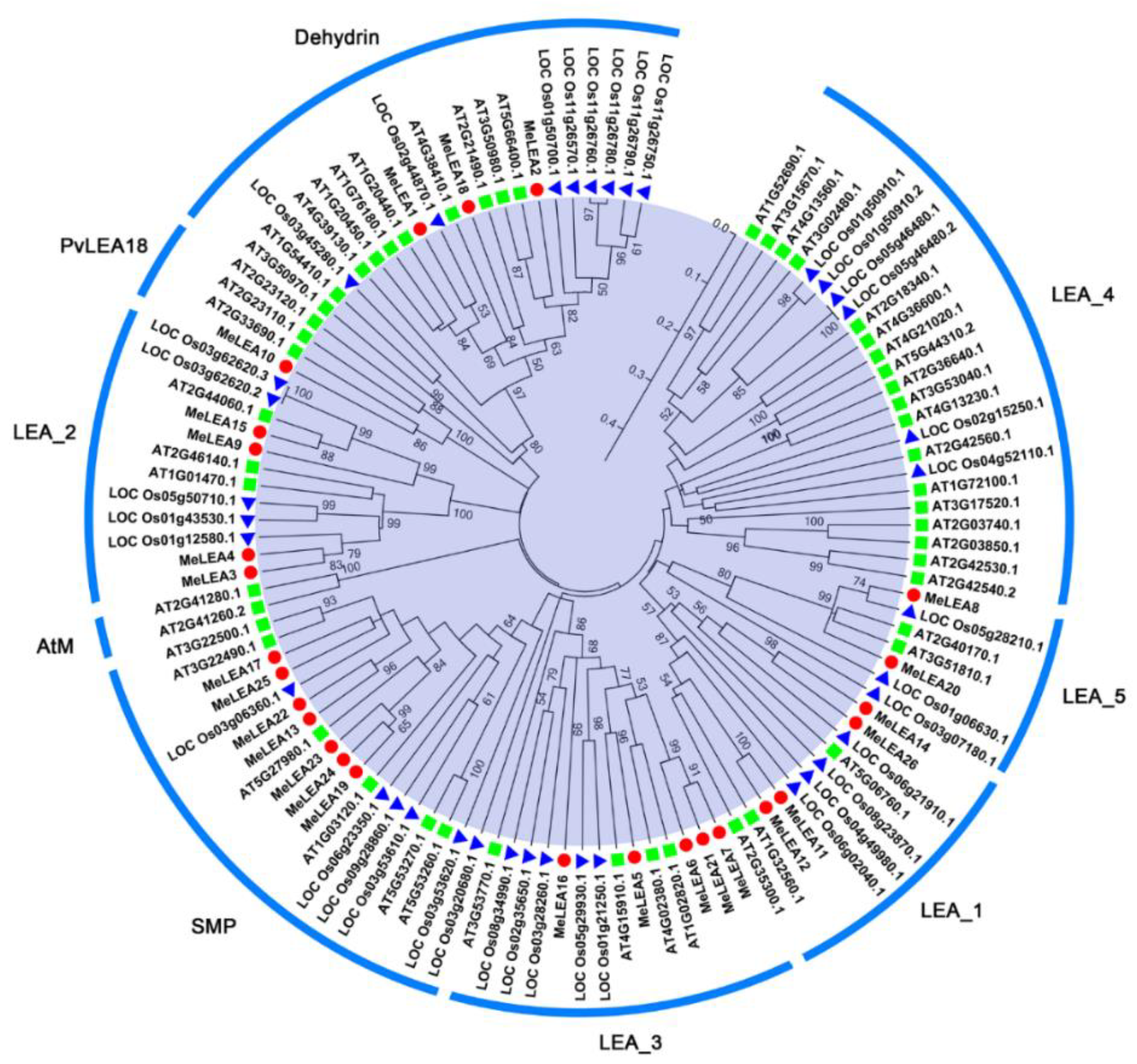
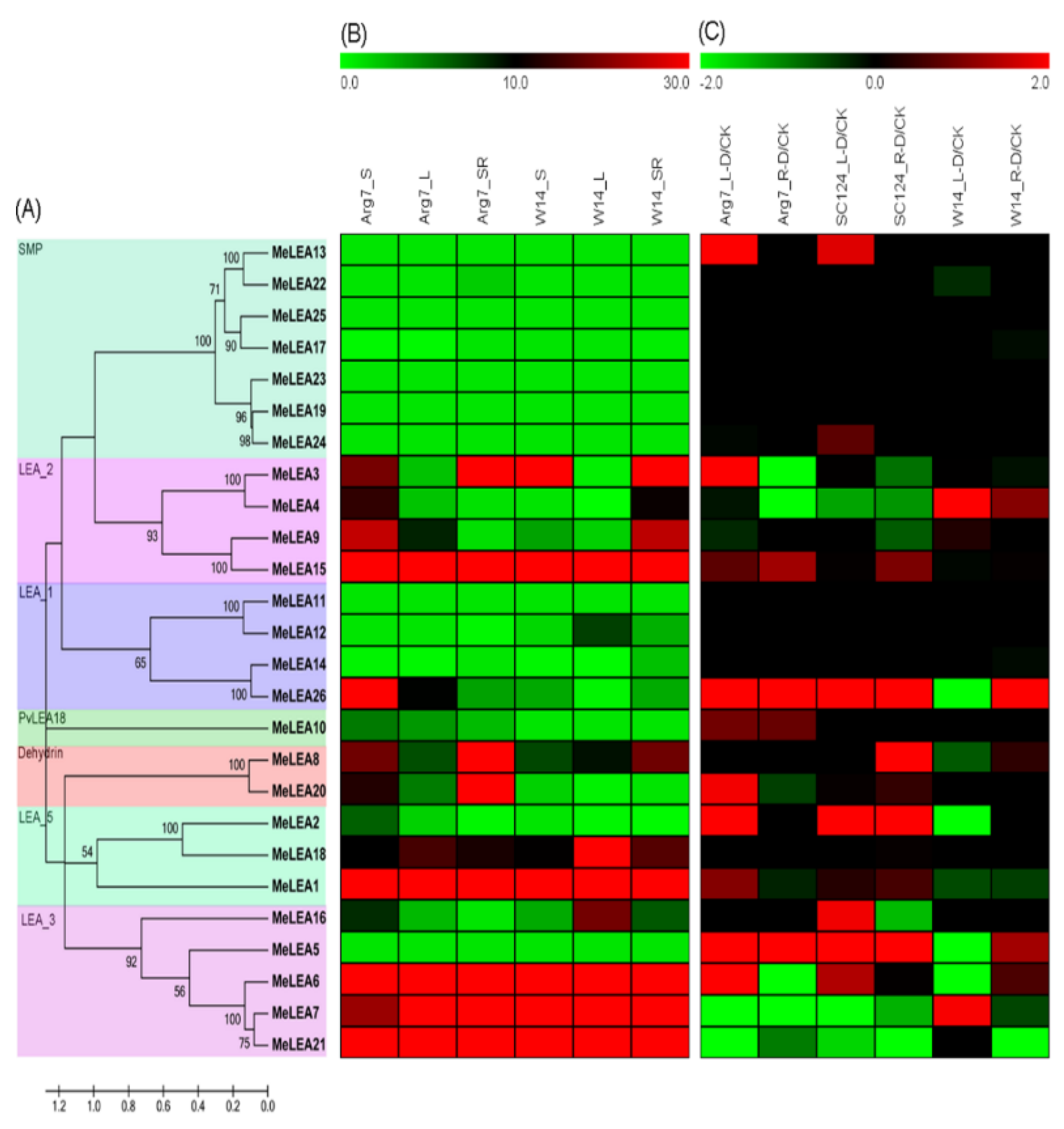
2.1. Identification and Phylogenetic Analysis of MeLEAs
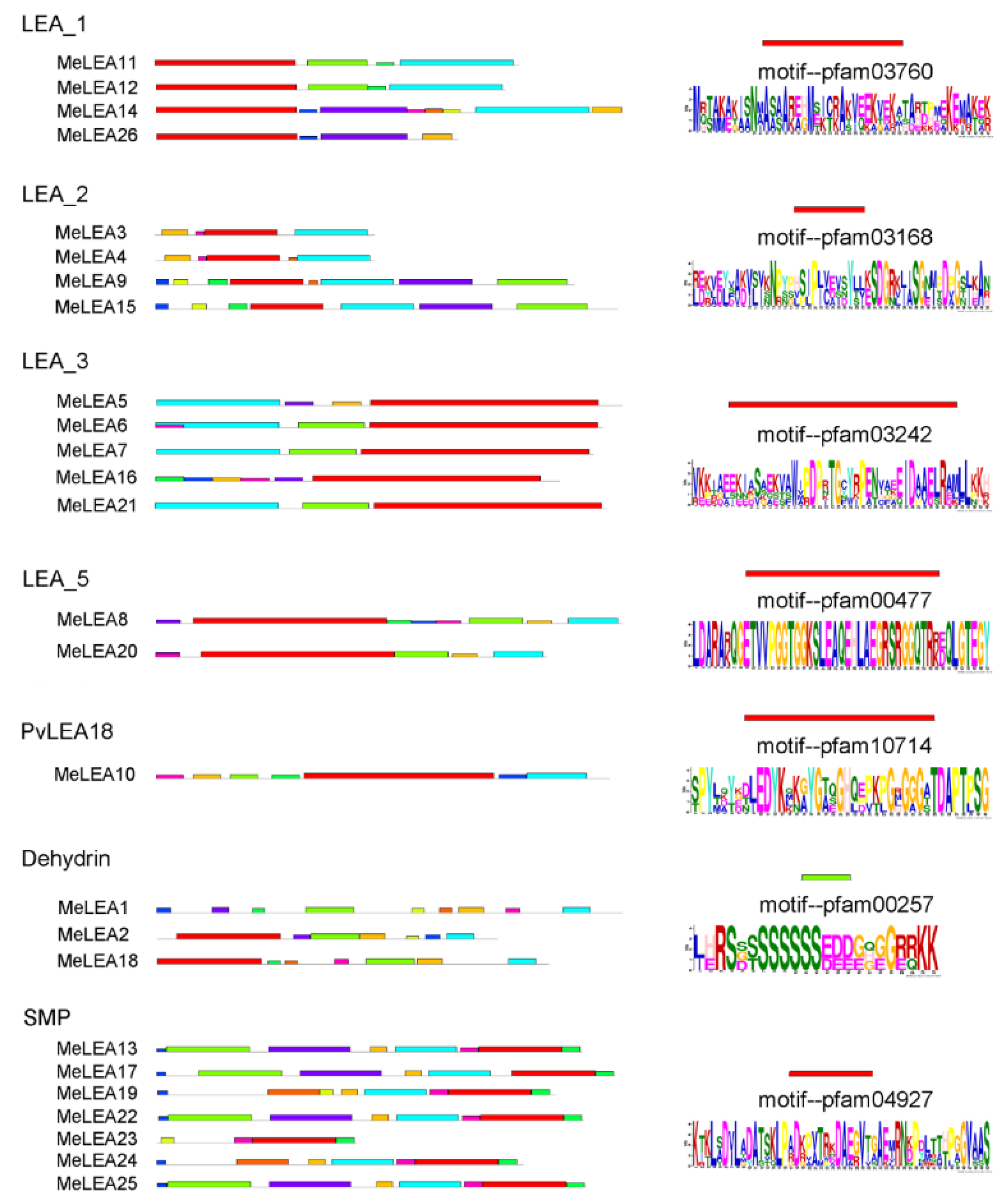
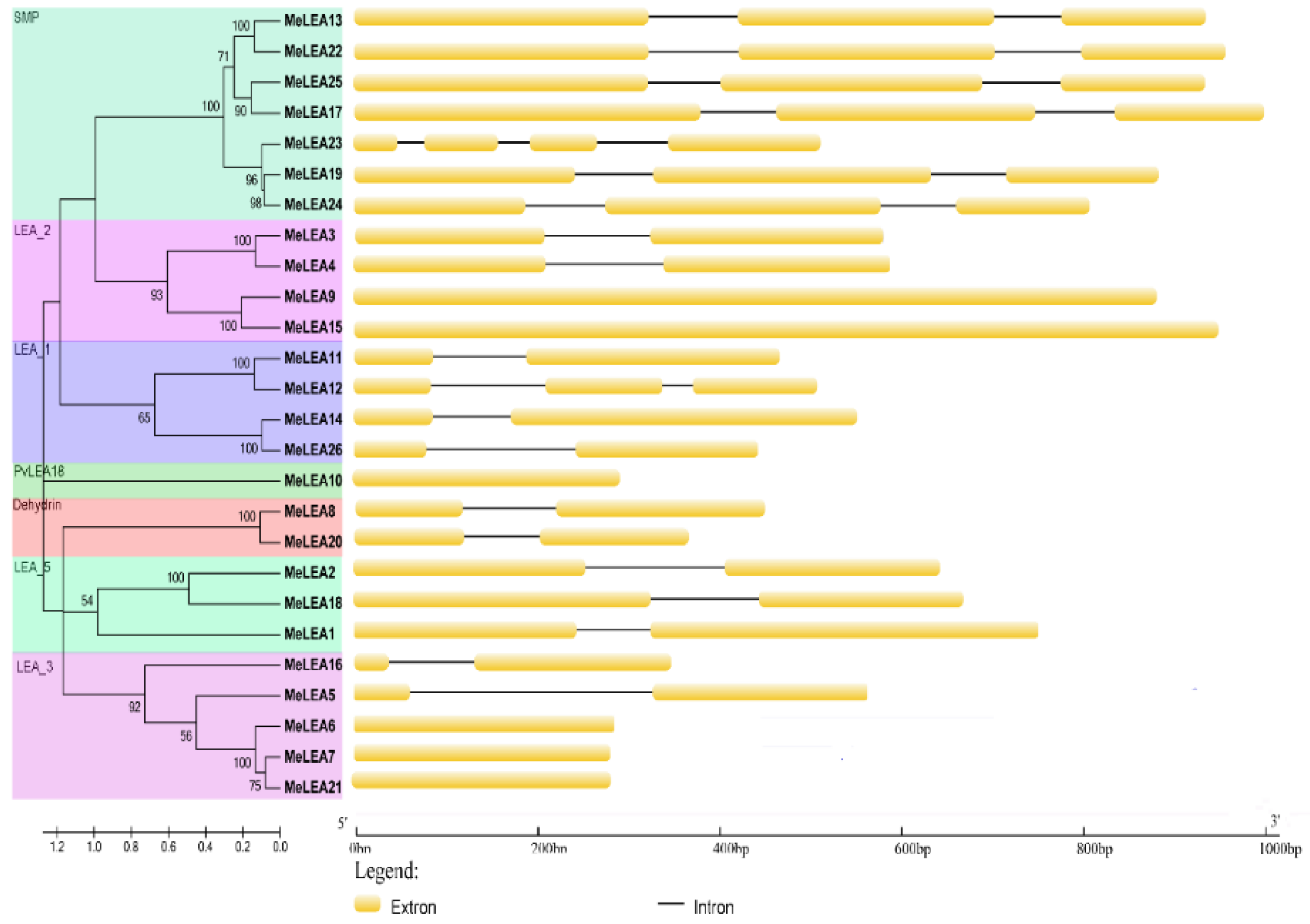
2.2. Conserved Motifs and Gene Structure of MeLEAs
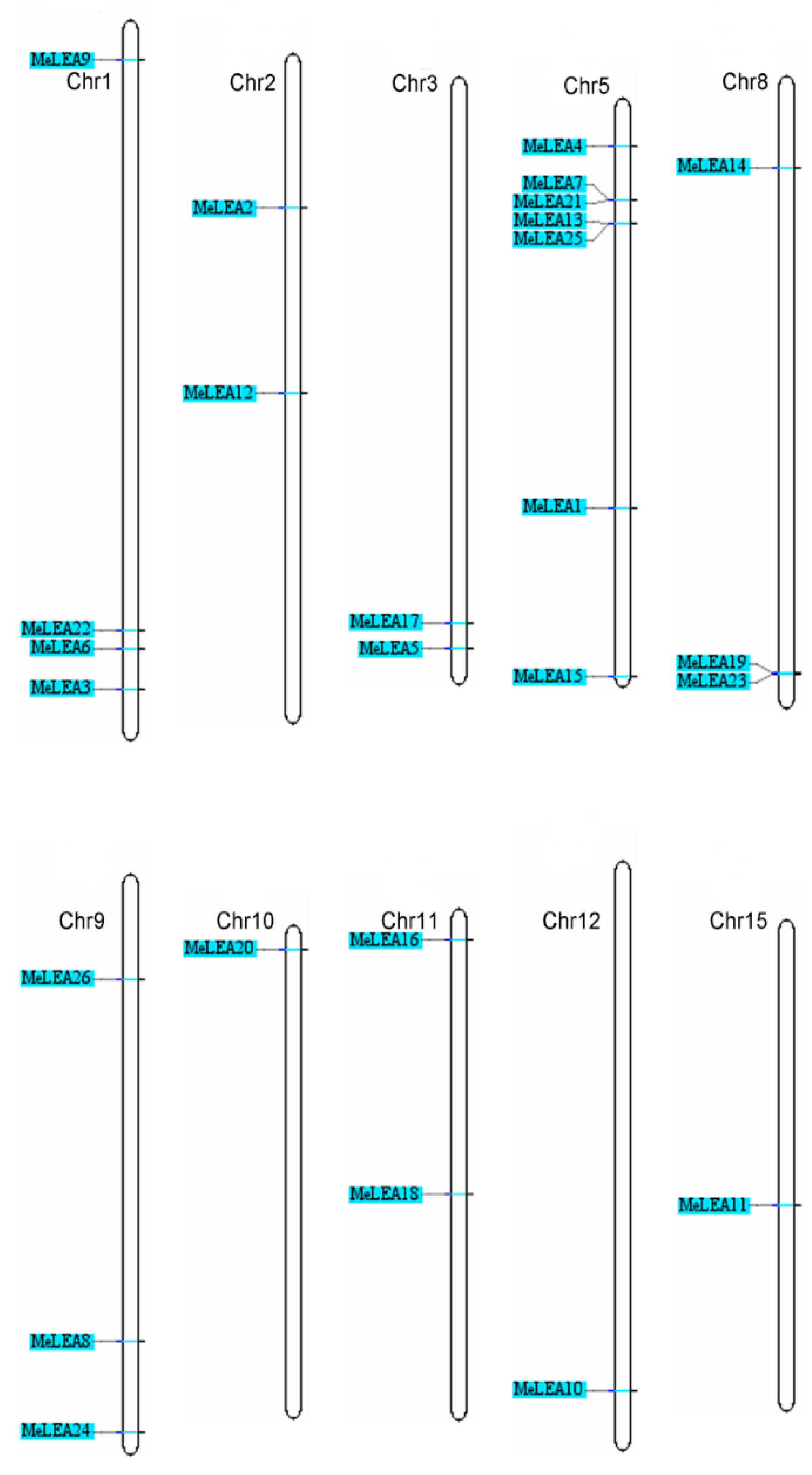
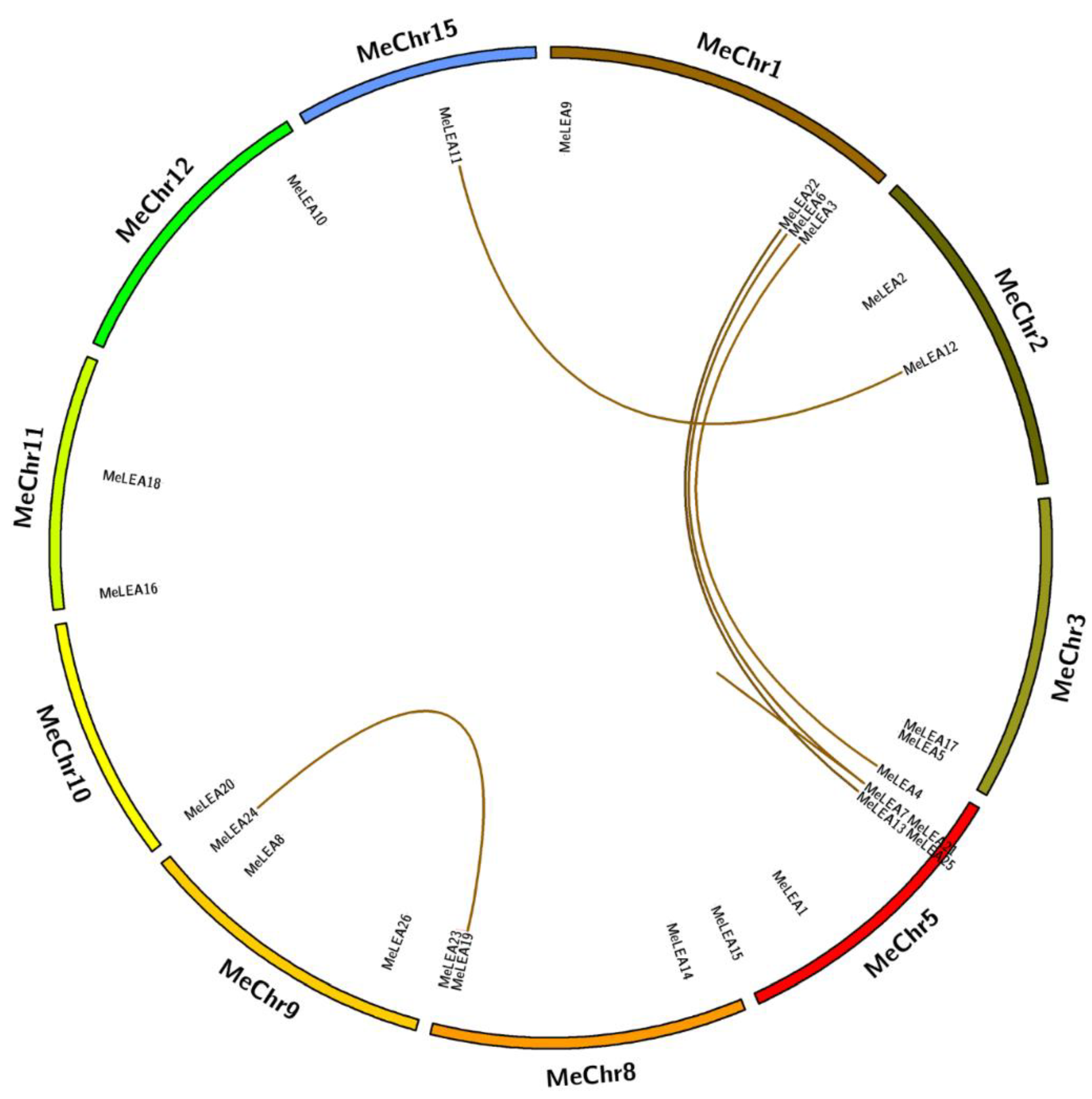
2.3. Chromosomal Distribution and Duplication Pattern of MeLEAs
2.4. Expression Profiles of LEA Genes in Different Cassava Tissues
2.5. Expression Profiles of LEA Genes in Response to Drought Stress
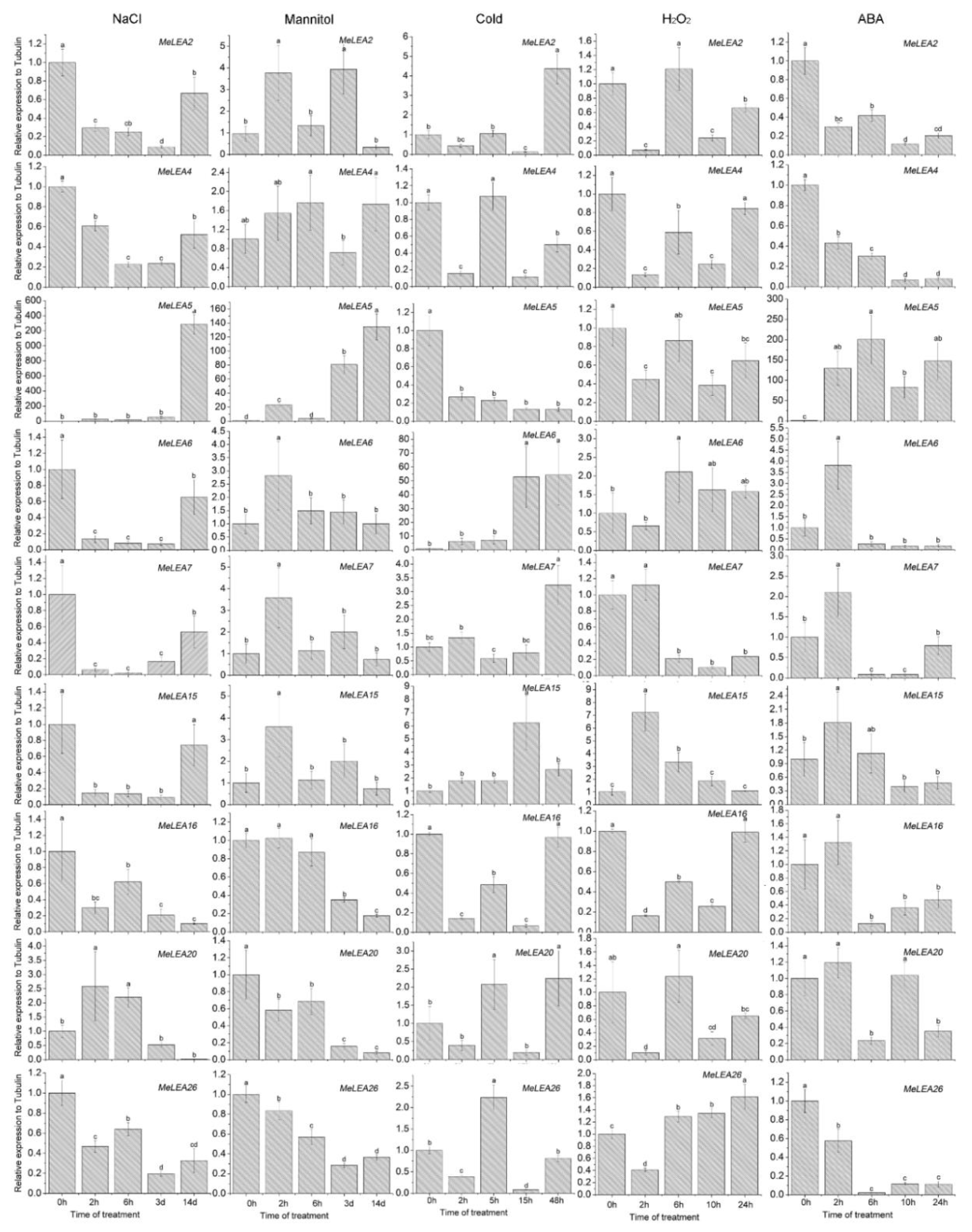
2.6. Expression Profiles of MeLEA Genes upon Exposure to Various Stresses and Related Signaling
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification and Phylogenetic Analyses
4.3. Protein Properties and Gene Structure Analysis
4.4. Chromosomal Location and Duplication Pattern Analysis
4.5. Transcriptomic Analysis
4.6. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dure, L.; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, H.; Jin, G.; Liu, H.; Wu, W.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Amara, I.; Odena, A.; Oliveira, E.; Moreno, A.; Masmoudi, K.; Pagès, M.; Goday, A. Insights into Maize LEA Proteins: From Proteomics to Functional Approaches. Plant Cell Physiol. 2012, 53, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W.; Zhu, B.; Close, T.J. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 1999, 98, 1234–1247. [Google Scholar] [CrossRef]
- Sasaki, K.; Christov, N.K.; Tsuda, S.; Imai, R. Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 2014, 55, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Prietodapena, P.; Almoguera, C.; Rojas, A.; Jordano, J. Seed-specific expression patterns and regulation by ABI3 of an unusual late embryogenesis-abundant gene in sunflower. Plant Mol. Biol. 1999, 39, 615–627. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Xiong, Z.; Zheng, J.; Xu, D.; Zhu, Z.; Xiang, J.; Gan, J.; Raboanatahiry, N.; Yin, Y.; Li, M. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 2016, 6, 24265. [Google Scholar] [CrossRef] [PubMed]
- Gal, T.Z.; Glazer, I.; Koltai, H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett. 2004, 577, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Kikawada, T.; Nakahara, Y.; Kanamori, Y.; Iwata, K.; Watanabe, M.; McGee, B.; Tunnacliffe, A.; Okuda, T. Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem. Biophys. Res. Commun. 2006, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Fu, J.; Stewart, M.; Ward, S.J.; Drew, J. The research progresses in Lea proteins of seeds. Plant Physiol. Commun. 1996, 32, 241–246. [Google Scholar]
- Garay-Arroyo, A.; Colmenero-Flores, J.M.; Garciarrubio, A.; Covarrubias, A.A. Highly Hydrophilic Proteins in Prokaryotes and Eukaryotes Are Common during Conditions of Water Deficit. J. Biol. Chem. 2000, 275, 5668–5674. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The Ubiquitous Distribution of Late Embryogenesis Abundant Proteins across Cell Compartments in Arabidopsis Offers Tailored Protection against Abiotic Stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyal, K.; Walton, L.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 2005, 388, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol. BioSyst. 2012, 8, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Krüger, C.; Berkowitz, O.; Stephan, U.W.; Hell, R.A. Metal-binding Member of the Late Embryogenesis Abundant Protein Family Transports Iron in the Phloem of Ricinus communis L. J. Biol. Chem. 2002, 277, 25062–25069. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Kanamori, Y.; Furuki, T.; Kikawada, T.; Okuda, T.; Takahashi, T.; Mihara, H.; Sakurai, M. Desiccation-induced structuralization and glass formation of group 3 late embryogenesis abundant protein model peptides. Biochemistry 2010, 49, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Cai, W. OsLEA3-2, an Abiotic Stress Induced Gene of Rice Plays a Key Role in Salt and Drought Tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, J.; Sun, L.; Yang, X.; Li, D. Group 3 LEA Protein, ZmLEA3, Is Involved in Protection from Low Temperature Stress. Front. Plant Sci. 2016, 7, 1011. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, W.; Xin, X.; Sun, L.P.; Pan, J.W.; Kong, X.P.; Zhang, M.Y.; Li, D.E. ZmLEA3, a Multifunctional Group 3 LEA Protein from Maize (Zea mays L.), is Involved in Biotic and Abiotic Stresses. Plant Cell Physiol. 2013, 54, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Houde, M.; Dallaire, S.; Dong, D.N.; Sarhan, F. Overexpression of the acidic dehydrin WCOR410 improves freezing tolerance in transgenic strawberry. Plant Biotechnol. J. 2004, 2, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, P.; Li, C.; Pan, Y.; Jiang, X.; Zhu, D.; Zhao, Q.; Yu, J. SiLEA14, a novel a typical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biol. 2014, 14, 290. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhou, M.; Zhou, X.; Jin, Y.; Xu, M.; Lin, J. JcLEA, a Novel LEA-Like Protein from Jatropha curcas, Confers a High Level of Tolerance to Dehydration and Salinity in Arabidopsis thaliana. PLoS ONE 2013, 8, e83056. [Google Scholar] [CrossRef] [PubMed]
- Dalal, M.; Tayal, D.; Chinnusamy, V.; Bansal, K.C. Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J. Biotechnol. 2009, 139, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhong, X.; He, J.; Jin, S.; Guo, H.; Yu, X.; Zhou, Y.; Li, X.; Ma, M.; Chen, Q.; et al. Genome-Wide Identification, Characterization, and Stress-Responsive Expression Profiling of Genes Encoding LEA (Late Embryogenesis Abundant) Proteins in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2016, 11, e0165953. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Ozyigit, I.I.; Tombuloglu, H.; Koc, I. In silico comparative analysis of lea (late embryogenesis abundant) proteins in brachypodium. Plant Omics 2013, 6, 433–440. [Google Scholar]
- Lan, T.; Gao, J.; Zeng, Q. Genome-wide analysis of the lea (late embryogenesis abundant) protein gene family in Populous trichocarpa. Tree Genet. Genomes 2013, 9, 253–264. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Santana, F.A.; Oliveira, L.A.; Santos, V.S. Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genet. Mol. Res. 2014, 13, 6683–6700. [Google Scholar] [CrossRef] [PubMed]
- Lebot, V.; Champagne, A.; Malapa, R.; Shiley, D. NIR Determination of Major Constituents in Tropical Root and Tuber Crop Flours. J. Agric. Food Chem. 2009, 57, 10539–10547. [Google Scholar] [CrossRef] [PubMed]
- Maran, J.P.; Sivakumar, V.; Thirugnanasambandham, K.; Sridhar, R. Degradation behavior of biocomposites based on cassava starch buried under indoor soil conditions. Carbohydr. Polym. 2014, 101, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Ding, Z.; Han, B.; Hu, W.; Li, Y.; Zhang, J. Physiological Investigation and Transcriptome Analysis of Polyethylene Glycol (PEG)-Induced Dehydration Stress in Cassava. Int. J. Mol. Sci. 2016, 17, 283. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Ding, Z.; Zhou, F.; Zhou, Y.; Yang, R.; Yang, Z.; Wang, W.; Peng, M. The Discrepant and Similar Responses of Genome-Wide Transcriptional Profiles between Drought and Cold Stresses in Cassava. Int. J. Mol. Sci. 2017, 18, 2668. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yu, X.; Lei, N.; Cheng, Z.; Zhao, P.; He, Y.; Wang, W.; Peng, M. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci. Rep. 2017, 7, 45981. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yan, Y.; Tie, W.; Ding, Z.; Wu, C.; Ding, X.; Wang, W.; Xia, Z.; Guo, J.; Peng, M. Genome-Wide Analyses of Calcium Sensors Reveal Their Involvement in Drought Stress Response and Storage Roots Deterioration after Harvest in Cassava. Genes 2018, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Yang, H.; Shi, H.; Wei, Y.; Tie, W.; Ding, Z.; Yan, Y.; Luo, Y.; Xia, Z.; Wang, W.; et al. The MAPKKK gene family in cassava: Genome-wide identification and expression analysis against drought stress. Sci. Rep. 2017, 7, 14939. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Fu, L.; Yan, Y.; Tie, W.; Xia, Z.; Wang, W.; Peng, M.; Hu, W.; Zhang, J. Genome-wide characterization and expression profiling of HD-Zip gene family related to abiotic stress in cassava. PLoS ONE 2017, 12, e0173043. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, A.M.; Martins, C.P.; Gonçalves, L.P.; Costa, M.G. Late Embryogenesis Abundant (LEA) Constitutes a Large and Diverse Family of Proteins Involved in Development and Abiotic Stress Responses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0145785. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, B.; Xiao, J.; Xia, Z.; Zhou, X.; Li, P.; Zhang, W.; Wang, Y.; Møller, B.L.; Zhang, P.; et al. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014, 5, 5110. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wei, Y.; Xia, Z.; Yan, Y.; Hou, X.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-Wide Identification and Expression Analysis of the NAC Transcription Factor Family in Cassava. PLoS ONE 2015, 10, e0136993. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Xia, Z.; Yan, Y.; Ding, Z.; Tie, W.; Wang, L.; Zou, M.; Wei, Y.; Lu, C.; Hou, X.; et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front. Plant Sci. 2015, 6, 914. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.C.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef] [PubMed]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 25 November 2017).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [PubMed]
- Brown, P.; Baxter, L.; Hickman, R.; Beynon, J.; Moore, J.D.; Ott, S. MEME-LaB: Motif analysis in clusters. Bioinformatics 2013, 29, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007, 396, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yue, H.; Feng, K.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.; Bouman, P.; Li, W. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Nekrutenko, A.; Makova, K.D.; Li, W. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Tang, H.; Tan, X.; Ficklin, S.P.; Feltus, F.A.; Paterson, A.H. Modes of Gene Duplication Contribute Differently to Genetic Novelty and Redundancy, but Show Parallels across Divergent Angiosperms. PLoS ONE 2011, 6, e28150. [Google Scholar] [CrossRef] [PubMed]
- FASTX-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools. Available online: http://hannonlab.cshl.edu/fastx_toolkit/index.html (accessed on 20 December 2017).
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 29 December 2017).
- Pollier, J.; Rombauts, S.; Goossens, A. Analysis of RNA-Seq data with TopHat and Cufflinks for genome-wide expression analysis of jasmonate-treated plants and plant cultures. Methods Mol. Biol. 2013, 1011, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Hu, W.; Yan, Y.; Tie, W.; Ding, Z.; Guo, J.; He, G. The Late Embryogenesis Abundant Protein Family in Cassava (Manihot esculenta Crantz): Genome-Wide Characterization and Expression during Abiotic Stress. Molecules 2018, 23, 1196. https://doi.org/10.3390/molecules23051196
Wu C, Hu W, Yan Y, Tie W, Ding Z, Guo J, He G. The Late Embryogenesis Abundant Protein Family in Cassava (Manihot esculenta Crantz): Genome-Wide Characterization and Expression during Abiotic Stress. Molecules. 2018; 23(5):1196. https://doi.org/10.3390/molecules23051196
Chicago/Turabian StyleWu, Chunlai, Wei Hu, Yan Yan, Weiwei Tie, Zehong Ding, Jianchun Guo, and Guangyuan He. 2018. "The Late Embryogenesis Abundant Protein Family in Cassava (Manihot esculenta Crantz): Genome-Wide Characterization and Expression during Abiotic Stress" Molecules 23, no. 5: 1196. https://doi.org/10.3390/molecules23051196



