Mitochondria Transcription Factor A: A Putative Target for the Effect of Melatonin on U87MG Malignant Glioma Cell Line
Abstract
:1. Introduction
2. Results
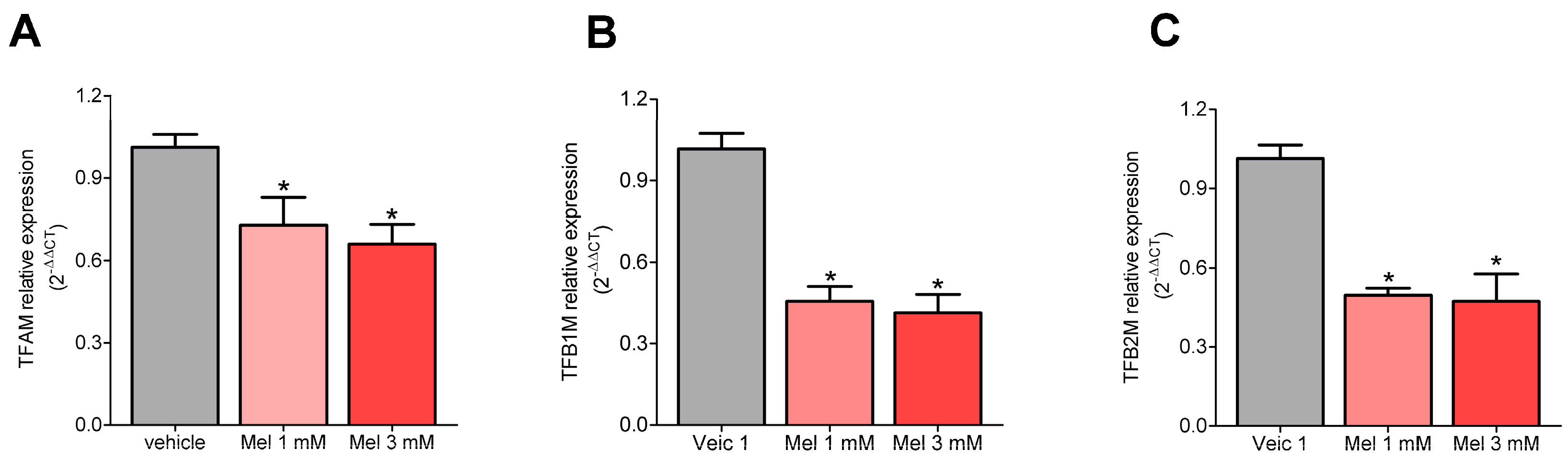
2.1. Melatonin Decreased the Expression of TFAM, TFB1M, and TFB2M
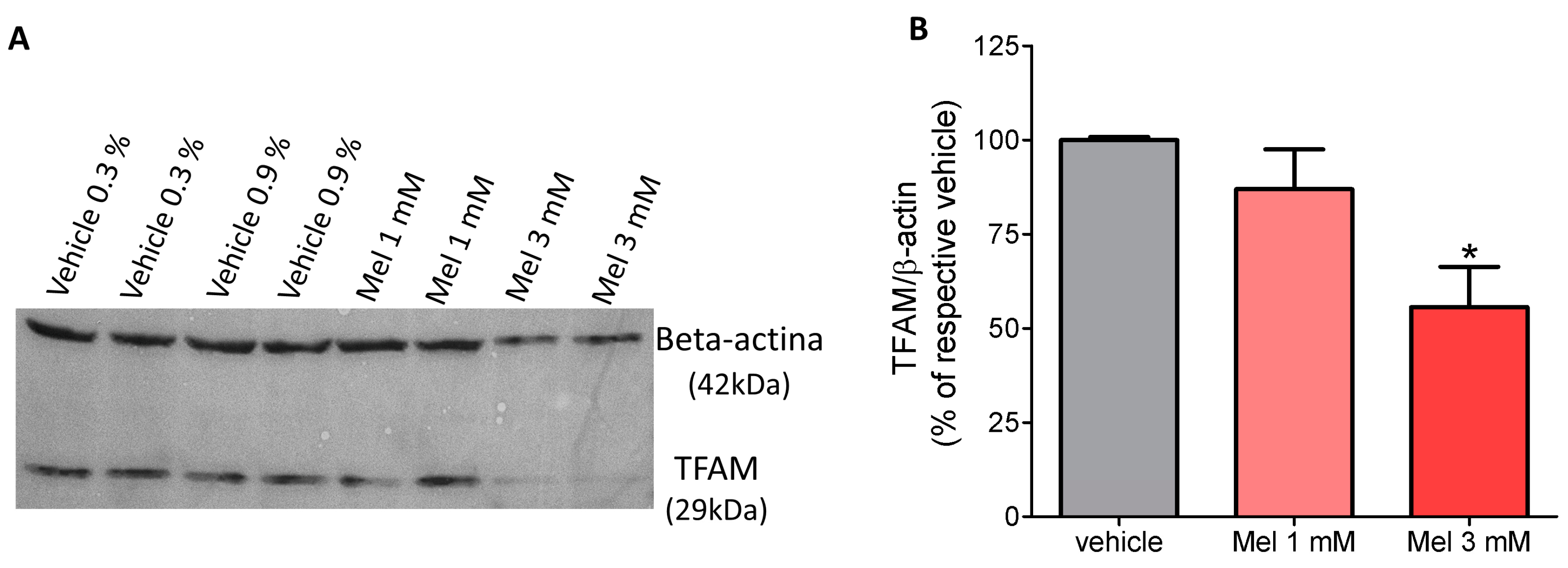
2.2. Melatonin Decreased the Content of TFAM Protein
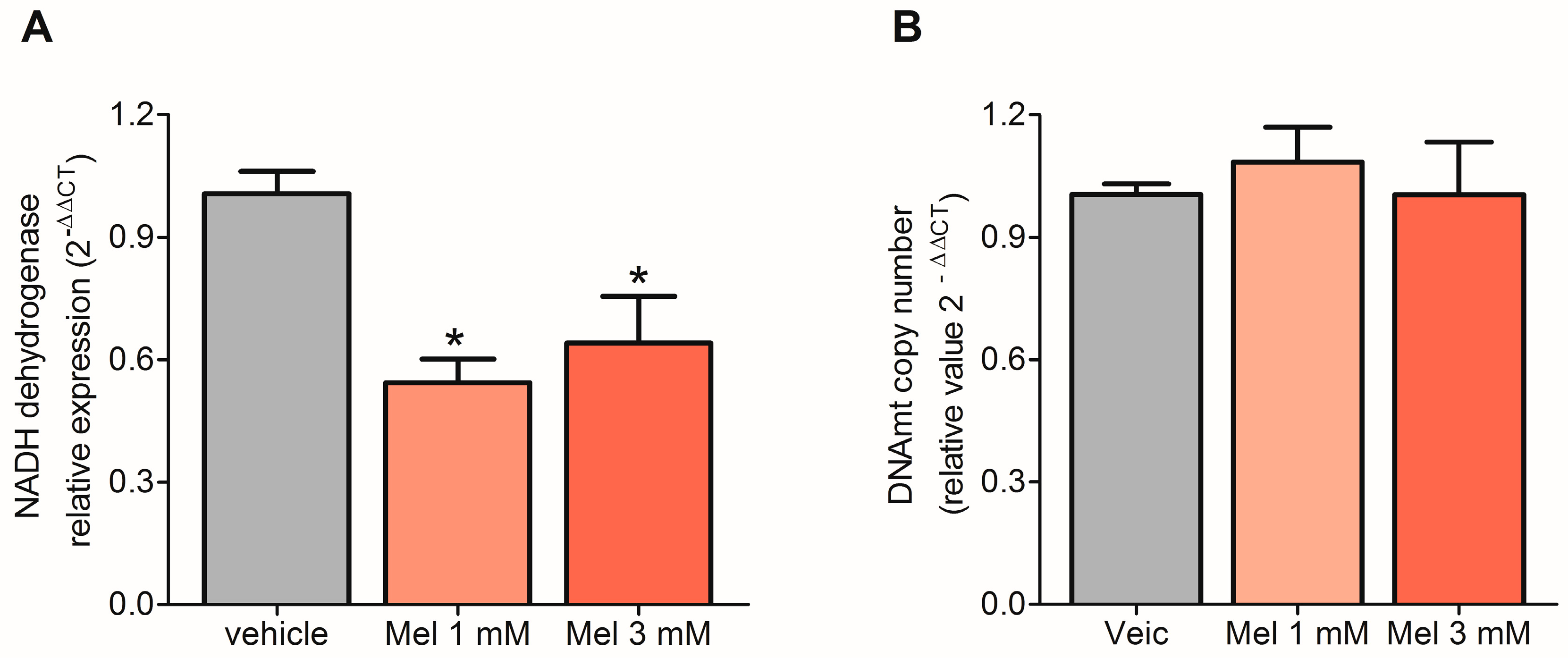
2.3. Melatonin Decreased the Transcription of mtDNA but Did Not Affect Replication
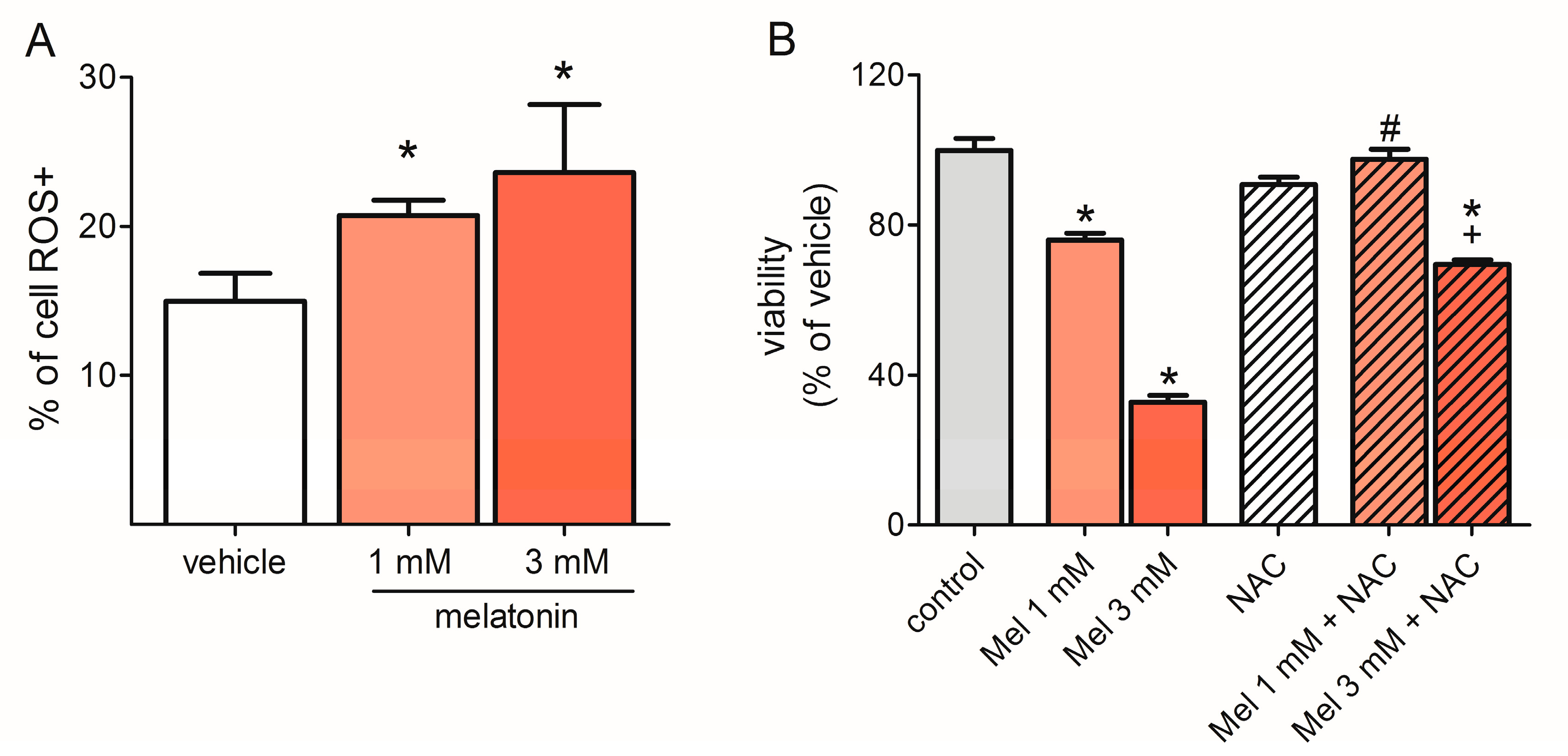
2.4. Melatonin Induced ROS Production
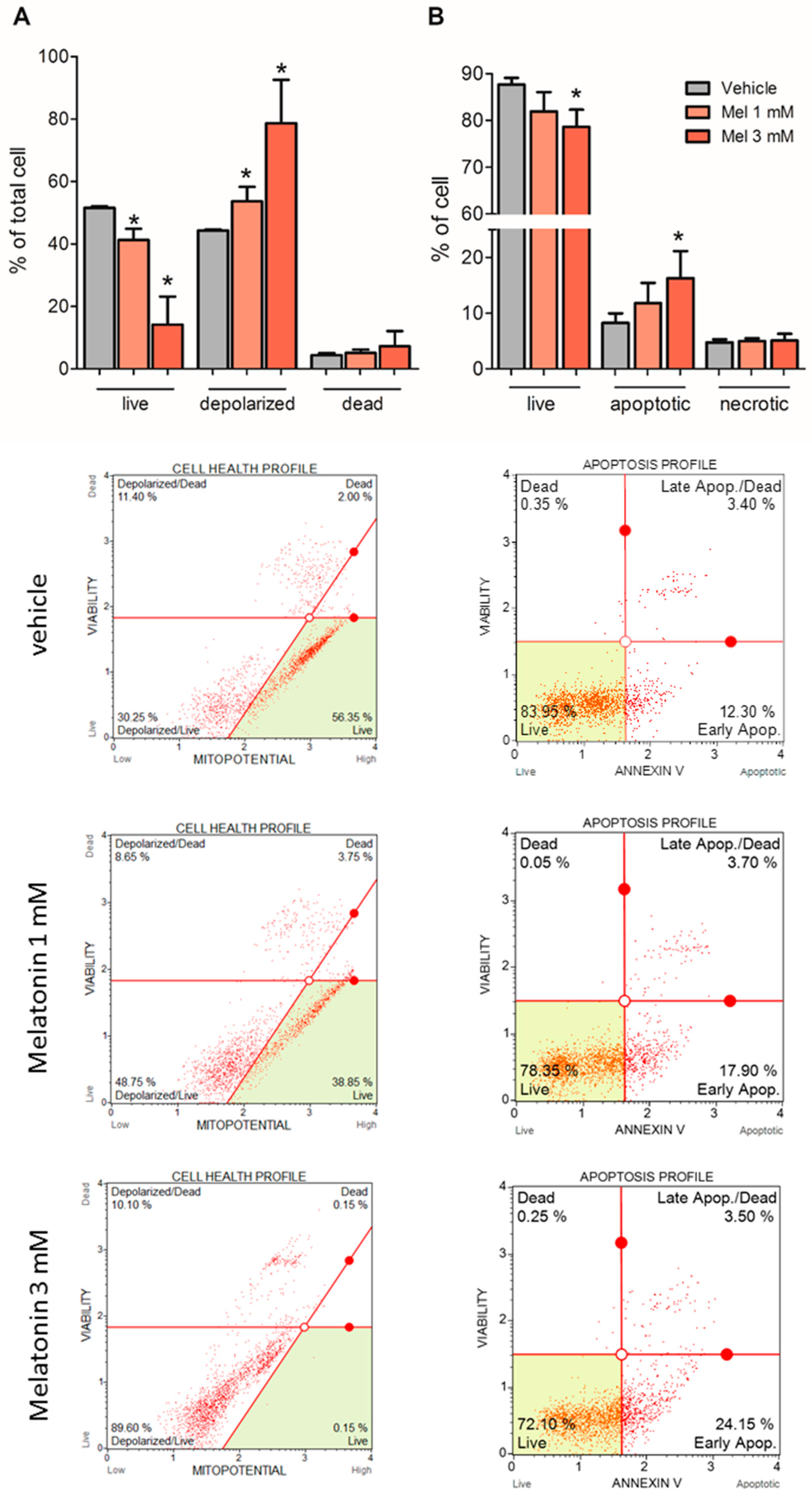
2.5. Melatonin Induced Mitochondria Depolarization and Apoptosis
2.6. Melatonin Arrested U87MG Cells at the G0/G1 Phase of the Cell Cycle
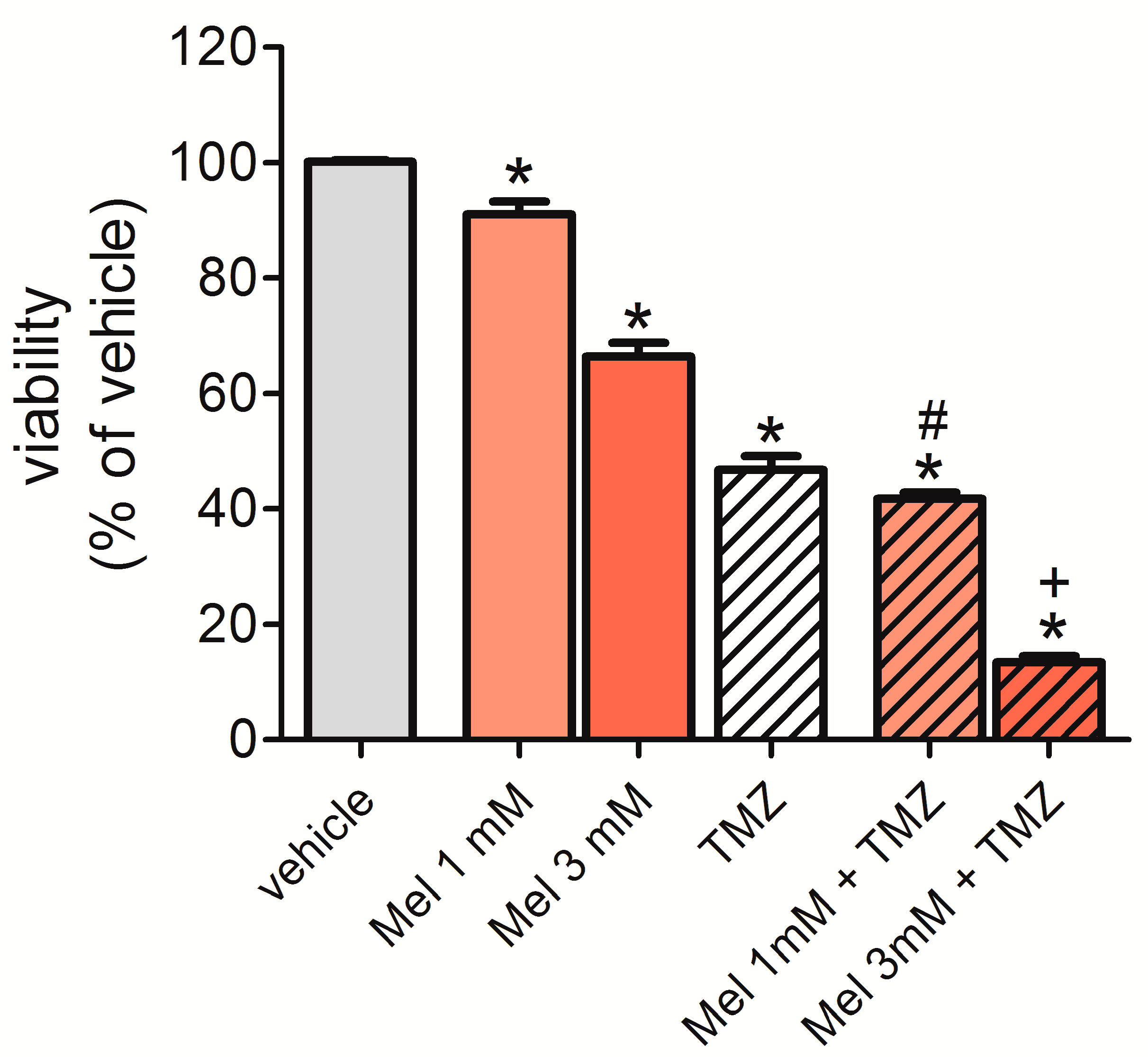
2.7. Melatonin Potentiated the Effect of Temozolomide (TMZ) to Reduce Cell Viability
3. Discussion
4. Materials and Methods
4.1. Cell Culture Conditions
4.2. Extraction of RNA and DNA
4.3. Expression of TFAM, TFB1M, TFB2M, and NADH Dehydrogenase 1 (MT-ND1) by qRT-PCR
4.4. Mitochondrial DNA Copy Number Quantification
4.5. Western Blot Analysis
4.6. Evaluation of Oxidative Stress, Cell Cycle, Apoptosis, and Mitochondria Polarization
4.7. Cell Viability/Proliferation
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nass, N.M. The circularity of Mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1966, 56, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Van Bruggen, E.F.; Borst, P.; Ruttenberg, G.J.; Gruber, M.; Kroon, A.M. Circular mitochondrial DNA. Biochim. Biophys. Acta 1966, 119, 437–439. [Google Scholar] [CrossRef]
- Kauppila, T.E.S.; Kauppila, J.H.K.; Larsson, N.G. Mammalian Mitochondria and Aging: An Update. Cell Metab. 2016, 25, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Baffy, G. Mitochondrial uncoupling in cancer cells: Liabilities andopportunities. Biochim. Biophys. Acta 2017, 185, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.P.; Chipuk, J.E. Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 2017, 74, 1999–2017. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, M.; Larsson, N.G.; Gustafsson, C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007, 76, 679–699. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta 2012, 1819, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.I.; Kanki, T.; Muta, T.; Ukaji, K.; Abe, Y.; Nakayama, H.; Takio, K.; Hamasaki, N.; Kang, D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003, 31, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Kanki, T.; Nakayama, H.; Sasaki, N.; Takio, K.; Alam, T.I.; Hamasaki, N.; Kang, D. Mitochondrial nucleoid and transcription factor A. Ann. N. Y. Acad. Sci. 2004, 1011, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.T.; Kolesar, J.E.; Kaufman, B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta 2012, 1819, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Ekstrand, M.I.; Falkenberg, M.; Rantanen, A.; Park, C.B.; Gaspari, M.; Hultenby, K.; Rustin, P.; Gustafsson, C.M.; Larsson, N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004, 13, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Lin, J.K.; Yang, S.H.; Wang, H.S.; Li, A.F.; Chang, S.C. Expression of beta-F1-ATPase and mitochondrial transcription factor A and the change in mitochondrial DNA content in colorectal cancer: Clinical data analysis and evidence from an in vitro study. Int. J. Colorectal. Dis. 2008, 23, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Correia, R.L.; Oba-Shinjo, S.M.; Uno, M.; Huang, N.; Marie, S.K. Mitochondrial DNA depletion and its correlation with TFAM, TFB1M, TFB2M and POLG in human diffusely infiltrating astrocytomas. Mitochondrion 2011, 11, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Hasegawa, J.; Nezu, R.; Kim, Y.K.; Hirota, M.; Kawano, K.; Izumi, H.; Kohno, K. Clinical usefulness of mitochondrial transcription factor A expression as a predictive marker in colorectal cancer patients treated with FOLFOX. Cancer Sci. 2011, 102, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yang, J.; Wang, Z.; Wu, G.; Liu, F. TFAM is directly regulated by miR-23b in glioma. Oncol. Rep. 2013, 30, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Mineo, J.F.; Bordron, A.; Baroncini, M.; Ramirez, C.; Maurage, C.A.; Blond, S.; Dam-Hieu, P. Prognosis factors of survival time in patients with glioblastoma multiforme: A multivariate analysis of 340 patients. Acta Neurochir. 2007, 149, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Furnari, F.B.; Fenton, T.; Bachoo, R.M.; Mukasa, A.; Stommel, J.M.; Stegh, A.; Hahn, W.C.; Ligon, K.L.; Louis, D.N.; Brennan, C.; et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007, 21, 2683–2710. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014, 9, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.R.; Correia, N.; Crossley, B.; Buckland, M.E.; Howell, V.M.; Wheeler, H.R. Correlation of MicroRNA 132 Up-regulation with an Unfavorable Clinical Outcome in Patients with Primary Glioblastoma Multiforme Treated with Radiotherapy Plus Concomitant and Adjuvant Temozolomide Chemotherapy. Transl. Oncol. 2013, 6, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Narita, Y.; Hawkins, C.E. Diffusely infiltrating astrocytomas: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 789–808. [Google Scholar] [CrossRef] [PubMed]
- Sainz, R.M.; Mayo, J.C.; Rodriguez, C.; Tan, D.X.; Lopez-Burillo, S.; Reiter, R.J. Melatonin and cell death: Differential actions on apoptosis in normal and cancer cells. Cell. Mol. Life Sci. 2003, 60, 1407–1426. [Google Scholar] [CrossRef] [PubMed]
- Luchetti, F.; Canonico, B.; Betti, M.; Arcangeletti, M.; Pilolli, F.; Piroddi, M.; Canesi, L.; Papa, S.; Galli, F. Melatonin signaling and cell protection function. FASEB J. 2010, 24, 3603–3624. [Google Scholar] [CrossRef] [PubMed]
- Radogna, F.; Nuccitelli, S.; Mengoni, F.; Ghibelli, L. Neuroprotection by melatonin on astrocytoma cell death. Ann. N. Y. Acad. Sci. 2009, 1171, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Lanoix, D.; Lacasse, A.A.; Reiter, R.J.; Vaillancourt, C. Melatonin: The smart killer: The human trophoblast as a model. Mol. Cell. Endocrinol. 2012, 348, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Acuña Castroviejo, D.; Escames, G.; Carazo, A.; León, J.; Khaldy, H.; Reiter, R.J. Melatonin, mitochondrial homeostasis and mitochondrial-related diseases. Curr. Top Med. Chem. 2002, 2, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Qin, L.; Reiter, R.J. Melatonin: A Mitochondrial Targeting Molecule Involving Mitochondrial Protection and Dynamics. Int. J. Mol. Sci. 2016, 17, 2124. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.; Macías, M.; Escames, G.; Reiter, R.J.; Agapito, M.T.; Ortiz, G.G.; Acuña-Castroviejo, D. Melatonin-induced increased activity of the respiratory chain complexes I and IV can prevent mitochondrial damage induced by ruthenium red in vivo. J. Pineal Res. 2000, 28, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Prunet-Marcassus, B.; Ambid, L.; Viguerie-Bascands, N.; Pénicaud, L.; Casteilla, L. Evidence for a direct effect of melatonin on mitochondrial genome expression of Siberian hamster brown adipocytes. J. Pineal Res. 2001, 30, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zheng, L.; Liu, W.; Wang, X.; Wang, Z.; Wang, Z.; French, A.J.; Kang, D.; Chen, L.; Thibodeau, S.N.; et al. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011, 71, 2978–2987. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, A.; Yeung, K.Y.; Donoghue, J.; Baker, M.J.; Kelly, R.D.; McKenzie, M.; Johns, T.G.; St. John, J.C. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 2013, 20, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Han, J. Mitochondrial DNA copy number alteration in human cancers. N. Am. J. Med. Sci. 2013, 6, 22–25. [Google Scholar] [CrossRef]
- Feng, Y.M.; Jia, Y.F.; Su, L.Y.; Wang, D.; Lv, L.; Xu, L.; Yao, Y.G. Decreased mitochondrial DNA copy number in the hippocampus and peripheral blood during opiate addiction is mediated by autophagy and can be salvaged by melatonin. Autophagy 2013, 9, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.C.; He, M.D.; Lu, Y.H.; Li, L.; Zhong, M.; Zhang, Y.W.; Wang, Y.; Yu, Z.P.; Zhou, Z. Nickel exposure induces oxidative damage to mitochondrial DNA in Neuro2a cells: The neuroprotective roles of melatonin. J. Pineal Res. 2011, 51, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef] [PubMed]
- De Miguel, M.; Cordero, M.D. Oxidative Therapy against Cancer. In Oxidative Stress and Diseases; Lushchak, V.I., Gospodaryov, D.V., Eds.; InTech: Rijeka, Croatia, 2012; Available online: http://www.intechopen.com/books/oxidative-stress-and-diseases/oxidative-therapy-against-cancer (accessed on 4 April 2018).
- Wu, C.C.; Bratton, S.B. Regulation of the intrinsic apoptosis pathway by reactive oxygen species. Antioxid. Redox Signal. 2013, 19, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, M.; Caffo, M.; Minutoli, L.; Marini, H.; Abbritti, R.V.; Squadrito, F.; Trichilo, V.; Valenti, A.; Barresi, V.; Altavilla, D.; et al. ROS and Brain Gliomas: An Overview of Potential and Innovative Therapeutic Strategies. Int. J. Mol. Sci. 2016, 17, 984. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Wu, X.; Lan, L.; Shangguan, F.; Lin, X.; Chen, F.; Xu, S.; Zhang, Y.; Chen, Z.; Huang, K.; et al. Downregulation of TFAM inhibits the tumorigenesis of non-small cell lung cancer by activating ROS-mediated JNK/p38MAPK signaling and reducing cellular bioenergetics. Oncotarget 2016, 7, 11609–11624. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, G.H.; Chaturvedi, P.; Tyagi, S.C. Mitochondrial pathways to cardiac recovery: TFAM. Heart Fail. Rev. 2016, 21, 499–517. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Yoshida, M.; Yamato, M.; Ide, T.; Wu, Z.; Ochi-Shindou, M.; Kanki, T.; Kang, D.; Sunagawa, K.; Tsutsui, H.; et al. Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor a in mice. J. Neurosci. 2008, 28, 8624–8634. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Ide, T.; Fujino, T.; Arai, S.; Saku, K.; Kakino, T.; Tyynismaa, H.; Yamasaki, T.; Yamada, K.; Kang, D.; et al. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS ONE 2015, 10, e0119687. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Gong, B.; Duan, W.; Fan, C.; Zhang, J.; Li, Z.; Xue, X.; Xu, Y.; Meng, D.; Li, B.; et al. Melatonin ameliorates myocardial ischemia/reperfusion injury in type 1 diabetic rats by preserving mitochondrial function: Role of AMPK-PGC-1α-SIRT3 signaling. Sci. Rep. 2017, 7, 41337. [Google Scholar] [CrossRef] [PubMed]
- Wölfler, A.; Caluba, H.C.; Abuja, P.M.; Dohr, G.; Schauenstein, K.; Liebmann, P.M. Prooxidant activity of melatonin promotes fas-induced cell death in human leukemic Jurkat cells. FEBS Lett. 2001, 502, 127–131. [Google Scholar] [CrossRef]
- Albertini, M.C.; Radogna, F.; Accorsi, A.; Uguccioni, F.; Paternoster, L.; Cerella, C.; De Nicola, M.; D’Alessio, M.; Bergamaschi, A.; Magrini, A.; et al. Intracellular pro-oxidant activity of melatonin deprives U937 cells of reduced glutathione without affecting glutathione peroxidase activity. Ann. N. Y. Acad. Sci. 2006, 1091, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Cristofanon, S.; Uguccioni, F.; Cerella, C.; Radogna, F.; Dicato, M.; Ghibelli, L.; Diederich, M. Intracellular prooxidant activity of melatonin induces a survival pathway involving NF-kappaB activation. Ann. N. Y. Acad. Sci. 2009, 1171, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Casado-Zapico, S.; Rodriguez-Blanco, J.; García-Santos, G.; Martín, V.; Sánchez-Sánchez, A.M.; Antolín, I.; Rodriguez, C. Synergistic antitumor effect of melatonin with several chemotherapeutic drugs on human Ewing sarcoma cancer cells: Potentiation of the extrinsic apoptotic pathway. J. Pineal Res. 2010, 48, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, I.; Espino, J.; Barriga, C.; Reiter, R.J.; Pariente, J.A.; Rodríguez, A.B. Pro-oxidant effect of melatonin in tumour leucocytes: Relation with its cytotoxic and pro-apoptotic effects. Basic Clin. Pharmacol. Toxicol. 2011, 108, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, I.; Espino, J.; Marchena, A.M.; Barriga, C.; Paredes, S.D.; Rodríguez, A.B.; Pariente, J.A. Melatonin enhances hydrogen peroxide-induced apoptosis in human promyelocytic leukaemia HL-60 cells. Mol. Cell. Biochem. 2011, 353, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Uguz, A.C.; Cig, B.; Espino, J.; Bejarano, I.; Naziroglu, M.; Rodríguez, A.B.; Pariente, J.A. Melatonin potentiates chemotherapy-induced cytotoxicity and apoptosis in rat pancreatic tumor cells. J. Pineal Res. 2012, 53, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Laothong, U.; Hiraku, Y.; Oikawa, S.; Intuyod, K.; Murata, M.; Pinlaor, S. Melatonin induces apoptosis in cholangiocarcinoma cell lines by activating the reactive oxygen species-mediated mitochondrial pathway. Oncol. Rep. 2015, 33, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Pariente, R.; Pariente, J.A.; Rodríguez, A.B.; Espino, J. Melatonin sensitizes human cervical cancer HeLa cells to cisplatin-induced cytotoxicity and apoptosis: Effects on oxidative stress and DNA fragmentation. J. Pineal Res. 2016, 60, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, H.; Yao, L.; Zhang, X.; Zhao, S.; Ling, E.A.; Hao, A.; Li, G. Melatonin suppresses migration and invasion via inhibition of oxidative stress pathway in glioma cells. J. Pineal Res. 2012, 53, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Martín, V.; Herrera, F.; Carrera-Gonzalez, P.; García-Santos, G.; Antolín, I.; Rodriguez-Blanco, J.; Rodriguez, C. Intracellular signaling pathways involved in the cell growth inhibition of glioma cells by melatonin. Cancer Res. 2006, 66, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.G.; Markus, R.P. The cellular state determines the effect of melatonin on the survival of mixed cerebellar cell culture. PLoS ONE 2014, 9, e106332. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P.; McStay, G.P.; Clarke, S.J. The permeability transition pore complex: Another view. Biochimie 2002, 84, 153–166. [Google Scholar] [CrossRef]
- León, J.; Acuña-Castroviejo, D.; Escames, G.; Tan, D.X.; Reiter, R.J. Melatonin mitigates mitochondrial malfunction. J. Pineal Res. 2005, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Andrabi, S.A.; Sayeed, I.; Siemen, D.; Wolf, G.; Horn, T.F. Direct inhibition of the mitochondrial permeability transition pore: A possible mechanism responsible for anti-apoptotic effects of melatonin. FASEB J. 2004, 18, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Izumi, H.; Torigoe, T.; Ishiguchi, H.; Itoh, H.; Kang, D.; Kohno, K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003, 63, 3729–3734. [Google Scholar] [PubMed]
- Wong, T.S.; Rajagopalan, S.; Freund, S.M.; Rutherford, T.J.; Andreeva, A.; Townsley, F.M.; Petrovich, M.; Fersht, A.R. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009, 37, 6765–6783. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Gao, J.; Zhang, L.; Zhou, H.; Fang, D.; Feng, S. P53 increase mitochondrial copy number via up-regulation of mitochondrial transcription factor A in colorectal cancer. Oncotarget 2016, 7, 75981–75995. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, M.A.; Liu, J.; Pelicano, H.; Hammoudi, N.; Croce, C.M.; Keating, M.J.; Huang, P. Alterations of mitochondrial biogenesis in chronic lymphocytic leukemia cells with loss of p53. Mitochondrion 2016, 31, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Antico Arciuch, V.G.; Elguero, M.E.; Poderoso, J.J.; Carreras, M.C. Mitochondrial regulation of cell cycle and proliferation. Antioxid. Redox Signal. 2012, 16, 1150–1180. [Google Scholar] [CrossRef]
- Gabrielson, M.; Björklund, M.; Carlson, J.; Shoshan, M. Expression of mitochondrial regulators PGC1α and TFAM as putative markers of subtype and chemoresistance in epithelial ovarian carcinoma. PLoS ONE 2014, 9, e107109. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.; Peng, F.; Wang, L.; Peng, L.; Lan, G.; Yu, S. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol. Lett. 2013, 6, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wu, M.H.; Wang, N.; Ying, M.Z.; Zhang, Y.Y.; Hua, J.; Chuan, L.; Wang, Y.J. Mitochondrial transcription factor A contributes to cisplatin resistance in patients with estrogen receptor-positive breast cancer. Mol. Med. Rep. 2016, 14, 5304–5310. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, Q.; Chen, N.; Sun, Y.; Wang, X.; Wu, L.; Chen, S.; Yuan, H.; Xu, A.; Wang, J. Mitochondrial transcription factor A regulated ionizing radiation-induced mitochondrial biogenesis in human lung adenocarcinoma A549 cells. J. Radiat. Res. 2013, 54, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhao, Z.; Xiao, Y.; Peng, J.; Chen, J.; He, Y. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of colon cancer. Mol. Med. Rep. 2016, 14, 5475–5480. [Google Scholar] [CrossRef] [PubMed]
- Kinker, G.S.; Oba-Shinjo, S.M.; Carvalho-Sousa, C.E.; Muxel, S.M.; Marie, S.K.; Markus, R.P.; Fernandes, P.A. Melatonergic system-based two-gene index is prognostic in human gliomas. J. Pineal Res. 2016, 60, 84–94. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not Available. |

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, D.G.; Moretti, I.F.; Marie, S.K.N. Mitochondria Transcription Factor A: A Putative Target for the Effect of Melatonin on U87MG Malignant Glioma Cell Line. Molecules 2018, 23, 1129. https://doi.org/10.3390/molecules23051129
Franco DG, Moretti IF, Marie SKN. Mitochondria Transcription Factor A: A Putative Target for the Effect of Melatonin on U87MG Malignant Glioma Cell Line. Molecules. 2018; 23(5):1129. https://doi.org/10.3390/molecules23051129
Chicago/Turabian StyleFranco, Daiane G., Isabele F. Moretti, and Suely K. N. Marie. 2018. "Mitochondria Transcription Factor A: A Putative Target for the Effect of Melatonin on U87MG Malignant Glioma Cell Line" Molecules 23, no. 5: 1129. https://doi.org/10.3390/molecules23051129





