New and Old Genes Associated with Primary and Established Responses to Paclitaxel Treatment in Ovarian Cancer Cell Lines
Abstract
:1. Introduction
2. Results
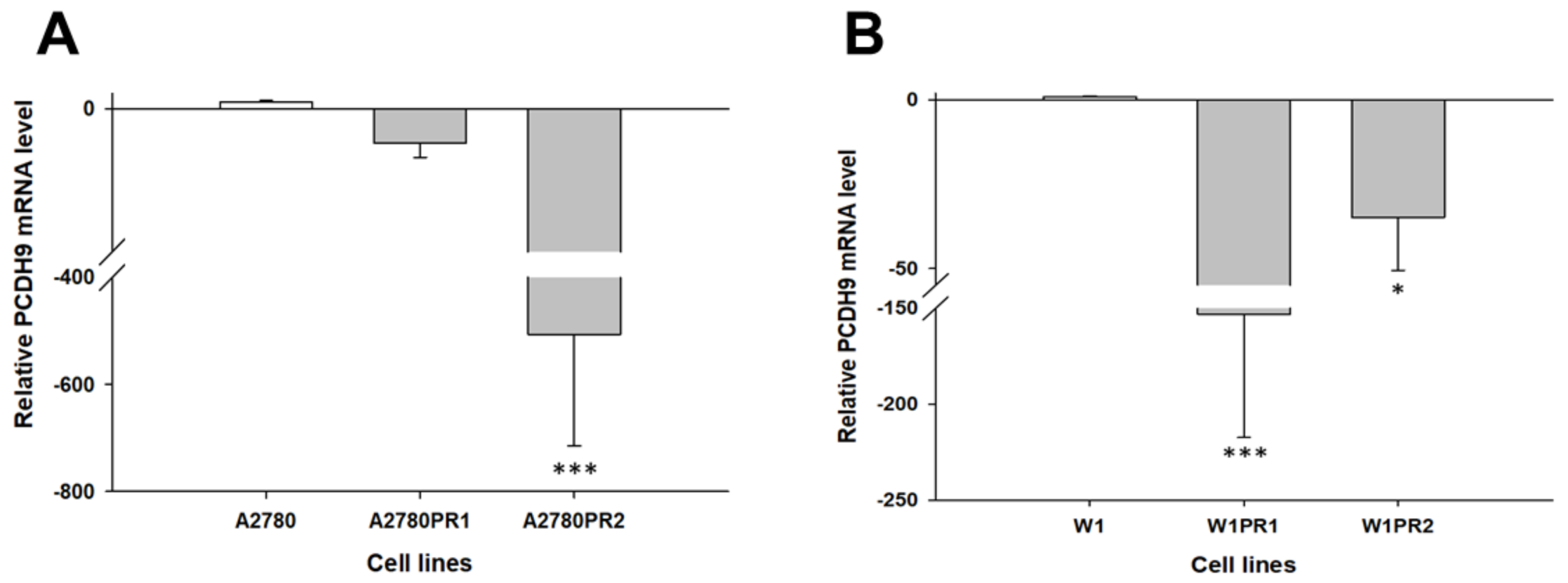
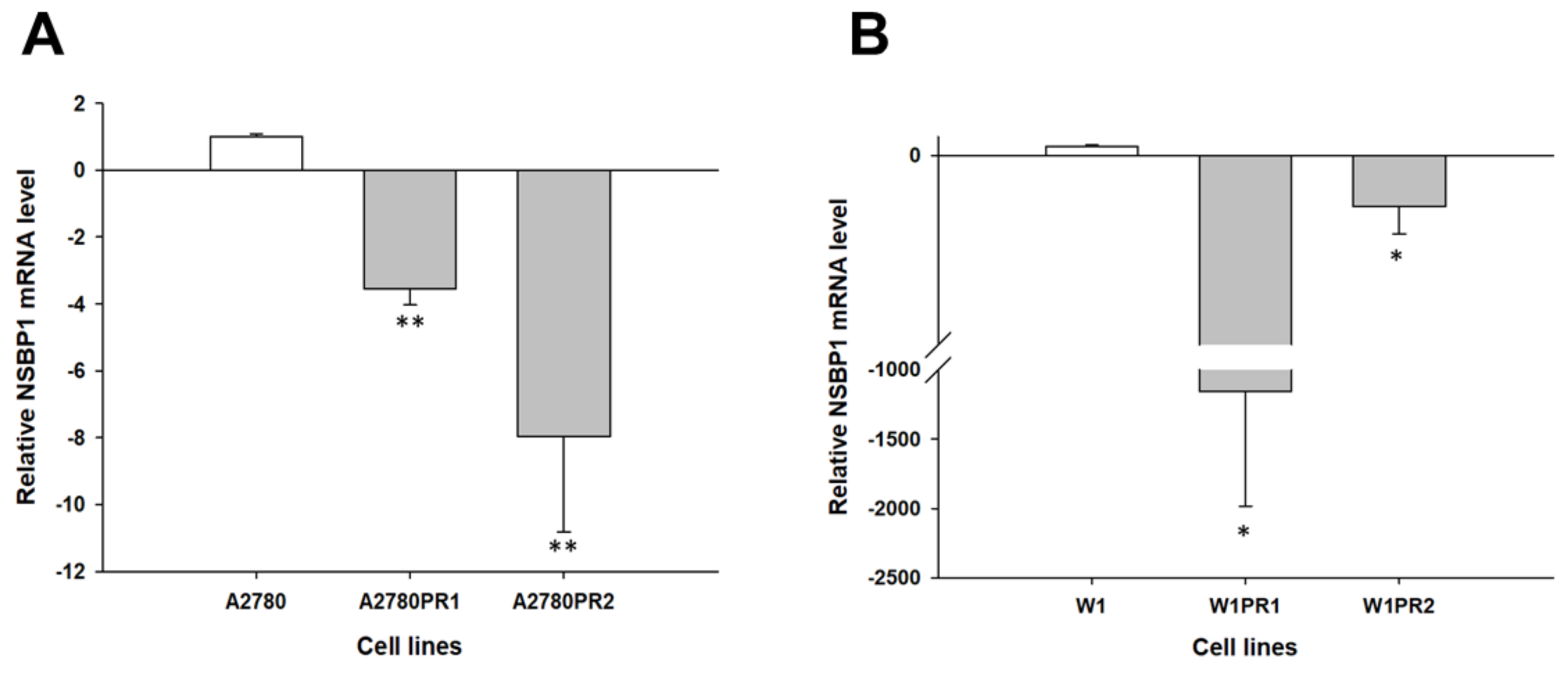
2.1. Gene Expression Analysis in PAC-Resistant Cell Lines
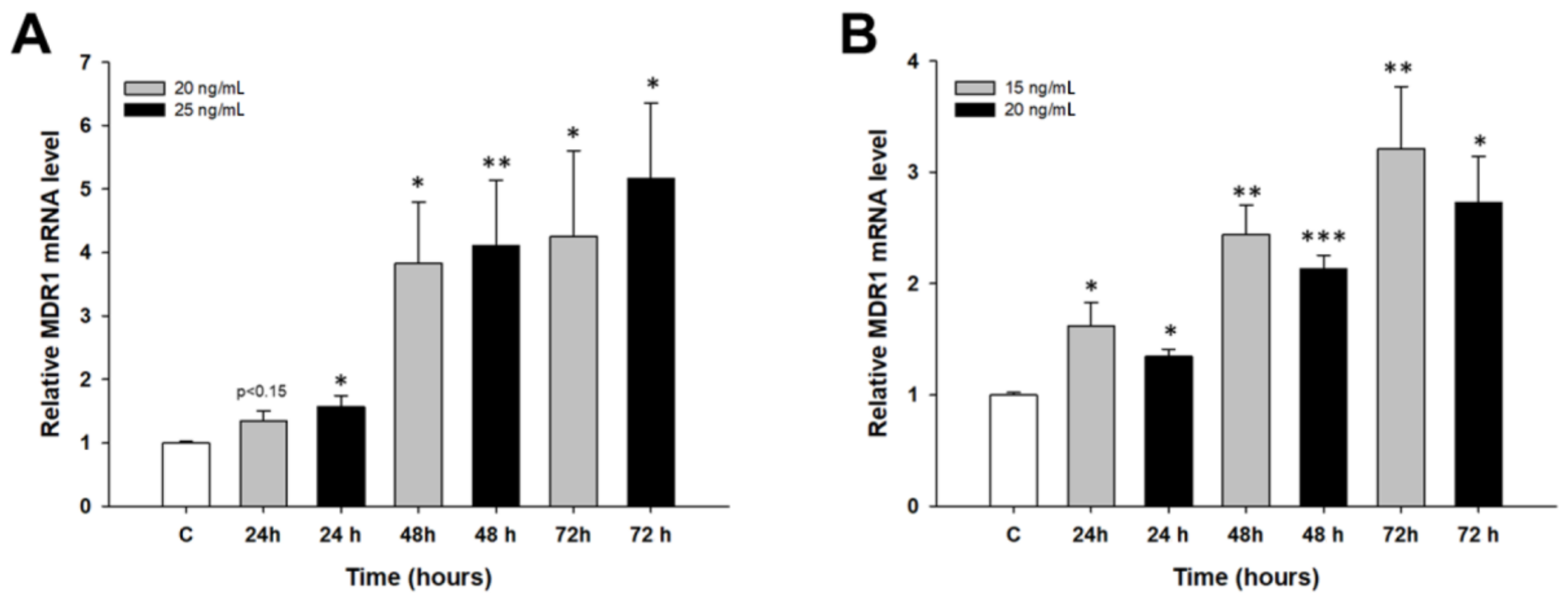
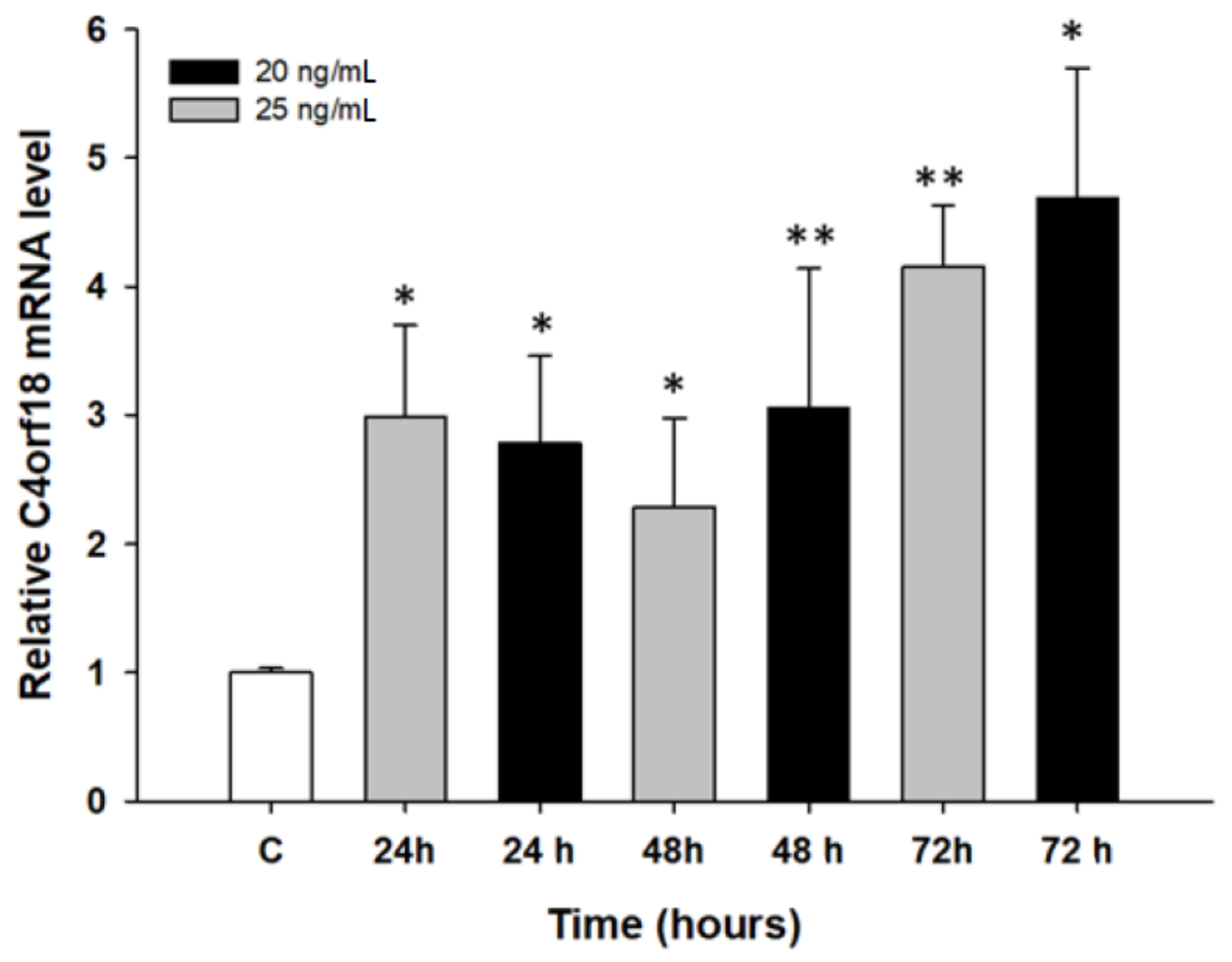
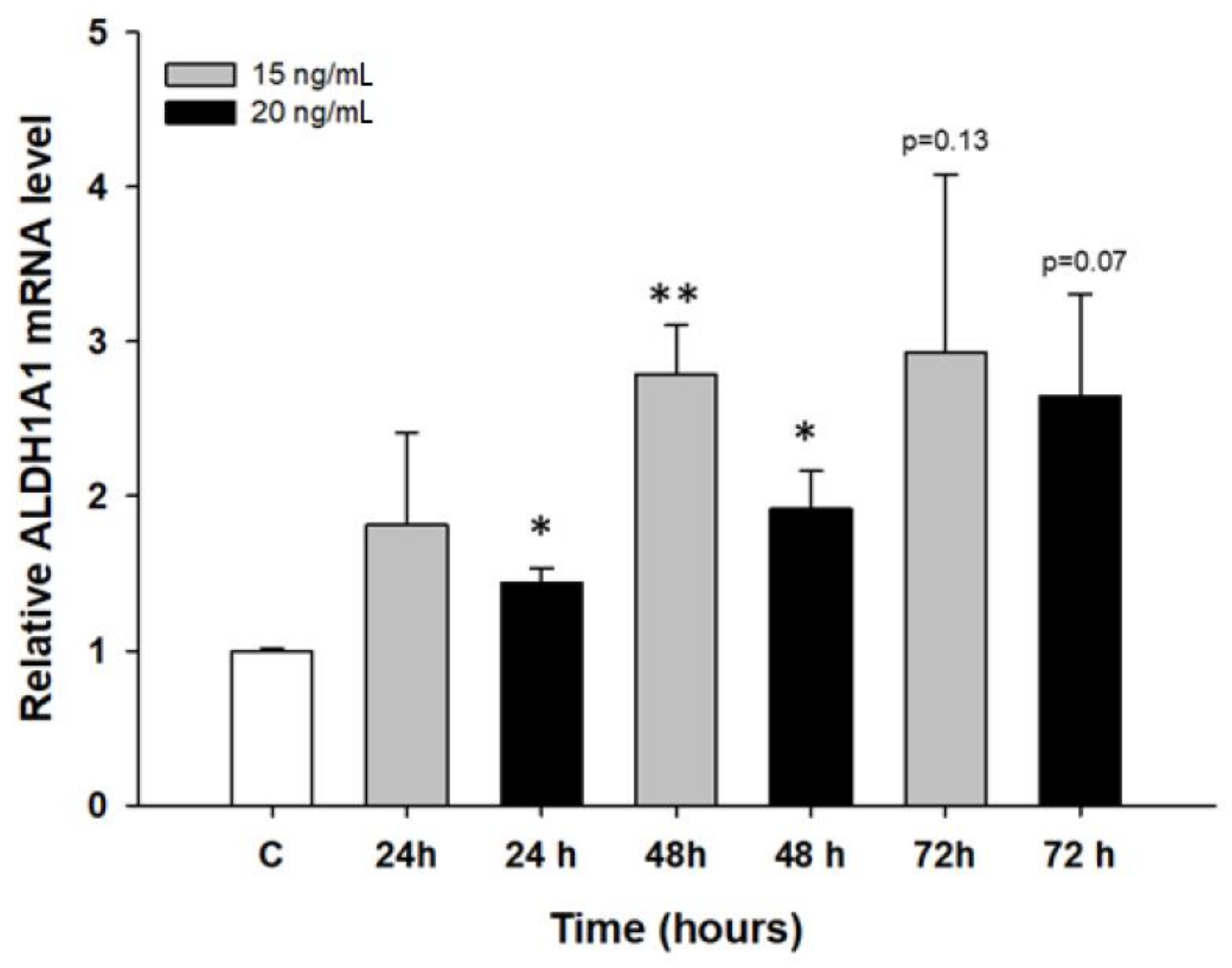
2.2. Early Response to PAC Treatment in Ovarian Cancer Cell Lines
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Lines and Cell Culture
4.3. Incubation of Cells with PAC
4.4. Examination of Gene Expression by Q-PCR
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2012, 62, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, B.T.; Coleman, R.L.; Markman, M. Ovarian cancer. Lancet 2009, 374, 1371–1382. [Google Scholar] [CrossRef]
- Parmar, M.K.; Ledermann, J.A.; Colombo, N.; du Bois, A.; Delaloye, J.F.; Kristensen, G.B.; Wheeler, S.; Swart, A.M.; Qian, W.; Torri, V.; et al. Paclitaxel plus platinum based chemotherapy versus conventional platinum-based chemo-therapy in women with relapsed ovarian cancer: The ICON4/AGO-OVAR-2.2 trial. Lancet 2003, 361, 2099–2106. [Google Scholar] [PubMed]
- Yu, S.; Li, A.; Liu, Q.; Yuan, X.; Xu, H.; Jiao, D.; Pestell, R.G.; Han, X.; Wu, K. Recent advances of bispecific antibodies in solid tumors. J. Hematol. Oncol. 2017, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Hadfield, J.A.; Lawrence, N.J.; McGown, A.T. Tubulin as a target for anticancer drugs: Agents which interact with the mitotic spindle. Med. Res. Rev. 1998, 18, 259–296. [Google Scholar] [CrossRef]
- McGuire, W.P.; Hoskins, W.J.; Brady, M.F.; Kucera, P.R.; Partridge, E.E.; Look, K.Y.; Clarke-Pearson, D.L.; Davidson, M. Cyclophosphamide and Cisplatin Compared with Paclitaxel and Cisplatin in Patients with Stage III and Stage IV Ovarian Cancer. N. Engl. J. Med. 1996, 334, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Holmes, F.A.; Walters, R.S.; Theriault, R.L.; Forman, A.D.; Newton, L.K.; Raber, M.N.; Buzdar, A.U.; Frye, D.K.; Hortobagyi, G.N. Phase II Trial of Taxol, an Active Drug in the Treatment of Metastatic Breast Cancer. J. Natl. Cancer Inst. 1991, 83, 1797–1805. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.K.; Fossella, F.V.; Winn, R.J.; Shin, D.M.; Hynes, H.E.; Gross, H.M.; Davilla, E.; Leimert, J.; Dhingra, H.; Raber, M.N.; et al. Phase II Study of Taxol in Patients with Untreated Advanced Non-Small-Cell Lung Cancer. J. Natl. Cancer Inst. 1993, 85, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Derry, W.B.; Wilson, L.; Khan, I.A.; Luduena, R.F.; Jordan, M.A. Taxol Differentially Modulates the Dynamics of Microtubules Assembled from Unfractionated and Purified Beta-Tubulin Isotypes. Biochemistry 1997, 36, 3554–3562. [Google Scholar] [CrossRef] [PubMed]
- Giannakakou, P.; Sackett, D.L.; Kang, Y.K.; Zhan, Z.; Buters, J.T.; Fojo, T.; Poruchynsky, M.S. Paclitaxel-Resistant Human Ovarian Cancer Cells Have Mutant Beta-Tubulins that Exhibit Impaired Paclitaxel-Driven Polymerization. J. Biol. Chem. 1997, 272, 17118–17125. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Sterzyńska, K.; Zaorska, K.; Sosińska, P.; Klejewski, A.; Brązert, M.; Nowicki, M.; Zabel, M. Analysis of MDR genes expression and cross-resistance in eight drug resistant ovarian cancer cell lines. J. Ovarian Res. 2016, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Wojtowicz, K.; Andrzejewska, M.; Zabel, M. Expression of MDR1 and MDR3 gene products in paclitaxel-, doxorubicin- and vincristine-resistant cell lines. Biomed. Pharmacother. 2014, 68, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Świerczewska, M.; Sterzyńska, K.; Wojtowicz, K.; Nowicki, M.; Zabel, M. Increased Expression of Several Collagen Genes is Associated with Drug Resistance in Ovarian Cancer Cell Lines. J. Cancer 2016, 7, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brązert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; Kemler, R. Protocadherins. Curr. Opin. Cell Biol. 2002, 14, 557–562. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yasuda, S.; Tanaka, H.; Yamagata, K.; Kim, H. Non-clustered protocadherin. Cell Adhes. Migr. 2011, 5, 97–105. [Google Scholar] [CrossRef]
- Wang, C.; Tao, B.; Li, S.; Li, B.; Wang, X.; Hu, G.; Li, W.; Yu, Y.; Lu, Y.; Liu, J. Characterizing the role of PCDH9 in the regulation of glioma cell apoptosis and invasion. J. Mol. Neurosci. 2014, 52, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, G.; Liu, J.; Wang, J.; Zhang, Y.; Zhang, X.; Zhou, Z.; Huang, Z. Downregulation of PCDH9 predicts prognosis for patients with glioma. J. Clin. Neurosci. 2012, 19, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiang, H.; Zhang, Y.; Wang, J.; Yu, G. Loss of PCDH9 is associated with the differentiation of tumor cells and metastasis and predicts poor survival in gastric cancer. Clin. Exp. Metast. 2015, 32, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Lv, J.; Yang, Z.; Guo, L.; Zhang, L.; Li, M.; Han, W.; Chen, X.; Zhuang, H.; Lu, F. Protocadherin 9 inhibits epithelial–mesenchymal transition and cell migration through activating GSK-3b in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2014, 452, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wei, G.H.; Liu, D.; Wang, L.; Hou, Y.; Zhu, S.; Peng, L.; Zhang, Q.; Cheng, Y.; Su, H.; et al. Whole-genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur. Urol. 2018, 73, 322–339. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhang, Z. Screening of potentially crucial genes and regulatory factors involved in epithelial ovarian cancer using microarray analysis. Oncol. Lett. 2017, 14, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Hock, R.; Furusawa, T.; Ueda, T.; Bustin, M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007, 17, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Rochman, M.; Malicet, C.; Bustin, M. HMGN5/NSBP1: A new member of the HMGN protein family that affects chromatin structure and function. Biochim. Biophys. Acta 2010, 1799, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.Q.; Yao, L.; Zhang, X.Y.; Li, X.S.; Zhou, L.Q. Knockdown of the nucleosome binding protein 1 inhibits the growth and invasion of clear cell renal cell carcinoma cells in vitro and in vivo. J. Exp. Clin. Cancer Res. 2012, 31, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Zhou, L.Q.; Yang, X.Z.; Ai, J.K.; Xin, D.Q.; Na, Y.Q.; Guo, Y.L. The NSBP1 expression is up-regulated in prostate cancer cell. Basic Med. Sci. Clin. 2004, 24, 393–397. [Google Scholar]
- Qu, J.; Yan, R.; Chen, J.; Xu, T.; Zhou, J.; Wang, M.; Chen, C.; Yan, Y.; Lu, Y. HMGN5: A potential oncogene in gliomas. J. Neurooncol. 2011, 104, 729–736. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liu, C.; Wang, B.; Li, N.; Zuo, G.; Gao, D. HMGN5 blockade by siRNA enhances apoptosis, suppresses invasion and increases chemosensitivity to temozolomide in meningiomas. Int. J. Oncol. 2015, 47, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, W.; Yan, Y.; Wu, S. Silencing HMGN5 suppresses cell growth and promotes chemosensitivity in esophageal squamous cell carcinoma. J. Biochem. Mol. Toxicol. 2017, 31, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shin, O.H.; Han, W.; Wang, Y.; Südhof, T.C. Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J. Biol. Chem. 2005, 280, 1641–1651. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Yu, H.; Liang, F. Multiple C2 domains transmembrane protein 1 is expressed in CNS neurons and possibly regulates cellular vesicle retrieval and oxidative stress. J. Neurochem. 2015, 135, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, W.M.; Lotsari-Salomaa, J.E.; Kaur, S.; Niskakoski, A.; Knuutila, S.; Järvinen, H.; Mecklin, J.P.; Peltomäki, P. The Role of Chromosomal Instability and Epigenetics in Colorectal Cancers Lacking β-Catenin/TCF Regulated Transcription. Gastroenterol. Res. Pract. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Comoglio, P.M.; Tamagnone, L.; Giordano, S. Invasive growth: A two-way street for semaphoring signalling. Nat. Cell Biol. 2004, 6, 1155–1157. [Google Scholar] [CrossRef] [PubMed]
- Maione, F.; Molla, F.; Meda, C.; Latini, R.; Zentilin, L.; Giacca, M.; Seano, G.; Serini, G.; Bussolino, F.; Giraudo, E. Semaphorin 3A is an endogenous angiogenesis inhibitor that blocks tumor growth and normalizes tumor vasculature in transgenic mouse models. J. Clin. Investig. 2009, 119, 3356–3372. [Google Scholar] [CrossRef] [PubMed]
- Bachelder, R.E.; Lipscomb, E.A.; Lin, X.; Wendt, M.A.; Chadborn, N.H.; Eickholt, B.J.; Mercurio, A.M. Competing autocrine pathways involving alternative neuropilin-1 ligands regulate chemotaxis of carcinoma cells. Cancer Res. 2003, 63, 5230–5233. [Google Scholar] [PubMed]
- Tang, C.; Gao, X.; Liu, H.; Jiang, T.; Zhai, X. Decreased expression of SEMA3A is associated with poor prognosis in gastric carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4782–4794. [Google Scholar] [PubMed]
- Jiang, H.; Qi, L.; Wang, F.; Sun, Z.; Huang, Z.; Xi, Q. Decreased semaphorin 3A expression is associated with a poor prognosis in patients with epithelial ovarian carcinoma. Int. J. Mol. Med. 2015, 35, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhang, W.; Zhang, Y.; Zhang, H.; Fu, Z.; Ye, J.; Liu, L.; Song, X.; Wu, Y. Expression of semaphorin 3A and neuropilin 1 with clinicopathological features and survival in human tongue cancer. Med. Oral Patol. Oral Cir. Bucal 2012, 17, e962–e968. [Google Scholar] [CrossRef] [PubMed]
- Świerczewska, M.; Klejewski, A.; Wojtowicz, K.; Brązert, M.; Iżycki, D.; Nowicki, M.; Zabel, M.; Januchowski, R. New and Old Genes Associated with Primary and Established Responses to Cisplatin and Topotecan Treatment in Ovarian Cancer Cell Lines. Molecules 2017, 22, 1717. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Wojtowicz, K.; Sterzyńska, K.; Sosińska, P.; Andrzejewska, M.; Zawierucha, P.; Nowicki, M.; Zabel, M. Inhibition of ALDH1A1 activity decreases expression of drug transporters and reduces chemotherapy resistance in ovarian cancer cell lines. Int. J. Biochem. Cell Biol. 2016, 78, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, X.; Li, X.; Xie, F.; Su, B.; Zhang, M.; Zhou, L. Expression of oncogenic HMGN5 increases the sensitivity of prostate cancer cells to gemcitabine. Oncol. Rep. 2015, 33, 1519–1525. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; He, L.; Gan, Y.; Zeng, Q.; Dai, Y.; Tan, J. MiR-186 suppresses the growth and metastasis of bladder cancer by targeting NSBP1. Diagn. Pathol. 2015, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Du, X.; Liu, A.; Li, P. Suppression of nucleosome-binding protein 1 by miR-326 impedes cell proliferation and invasion in non-small cell lung cancer cells. Oncol. Rep. 2016, 35, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zhou, L.Q.; Zhang, X.Y. Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can inhibit the in vitro and in vivo proliferation of prostate cancer cells. Asian J. Androl. 2010, 12, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Huang, J.H.; Liu, W. Sema3A drastically suppresses tumor growth in oral cancer Xenograft model of mice. BMC Pharmacol. Toxicol. 2017, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.I.; Haber, M.; Henderson, M.J.; Norris, M.D. ABC transporters in cancer: more than just drug efflux pumps. Nat. Rev. Cancer. 2010, 10, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Maliepaard, M.; van Gastelen, M.A.; de Jong, L.A.; Pluim, D.; van Waardenburg, R.C.; Ruevekamp-Helmers, M.C.; Floot, B.G.; Schellens, J.H. Overexpression of the BCRP/MXR/ABCP gene in a topotecan selected ovarian tumor cell line. Cancer Res. 1999, 59, 4559–4563. [Google Scholar] [PubMed]
- Januchowski, R.; Wojtowicz, K.; Sujka-Kordowska, P.; Andrzejewska, M.; Zabel, M. MDR gene expression analysis of six drug-resistant ovarian cancer cell lines. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Klejewski, A.; Świerczewska, M.; Zaorska, K.; Brązert, M.; Nowicki, M.; Zabel, M.; Januchowski, R. New and Old Genes Associated with Topotecan Resistance Development in Ovarian Cancer Cell Lines. Anticancer Res. 2017, 37, 1625–1636. [Google Scholar] [PubMed]
- Januchowski, R.; Wojtowicz, K.; Zabel, M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013, 67, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wei, X.; Lin, S.; Qin, L.; Cheng, L.; Li, P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Qiang, W.; Guo, H.; Xu, X.; Kim, J.J.; Mazar, A.; Kong, B.; Wei, J.J. Histologic and molecular analysis of patient derived xenografts of high-grade serous ovarian carcinoma. J. Hematol. Oncol. 2016, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Correia, A.L.; Bissell, M.J. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist. Updat. 2012, 15, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gilligan, B.M.; Yuan, J.; Li, T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J. Hematol. Oncol. 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Shengnan, Y.; Anping, L.; Qian, L.; Tengfei, L.; Xun, Y.; Xinwei, H.; Kongming, W. Chimeric antigen receptor T cells: A novel therapy for solid tumors. J. Hematol. Oncol. 2017, 10, 78. [Google Scholar]
- Zhang, L.; Han, J.; Jackson, A.L.; Clark, L.N.; Kilgore, J.; Guo, H.; Livingston, N.; Batchelor, K.; Yin, Y.; Gilliam, T.P.; et al. NT1014, a novel biguanide, inhibits ovarian cancer growth in vitro and in vivo. J. Hematol. Oncol. 2016, 9, 91. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |




| Transcript | Sequence (5′–3′ Direction) | ENST Number | Product Size | |
|---|---|---|---|---|
| MCTP1 | F | AGAACCTCAACCCTGTGTGG | 00000312216 | 123 bp |
| R | AGGCTGAGCCCATAAAGTCA | |||
| PCDH9 | F | CTCTCCGGACAAGAGGACTG | 00000544246 | 125 bp |
| R | AGTGACCCAAAACCAAGCAC | |||
| NSBP1 | F | TGTGCCAGTTACACCAGAGG | 00000358130 | 124 bp |
| R | TTGCTTGGTTTCAGCAACTG | |||
| C4orf18 | F | GAGTACCCAAGCCTGAATCG | 00000393807 | 137 bp |
| R | ATCTTCCTTGCGAGGTCTGA | |||
| SEMA3A | F | TGTTGGAGCAAAGGATCACA | 00000265362 | 109 bp |
| R | AGCCCACTTGCATTCATCTC | |||
| ALDH1A1 | F | GTTGTCAAACCAGCAGAGCA | 00000165092 | 115 bp |
| R | CTGTAGGCCCATAACCAGGA | |||
| MDR1 | F | TGACAGCTACAGCACGGAAG | 00000265724 | 131 bp |
| R | TCTTCACCTCCAGGCTCAGT | |||
| BCRP | F | TTCGGCTTGCAACAACTATG | 00000237612 | 128 bp |
| R | TCCAGACACACCACGGATAA | |||
| GAPDH | F | GAAGGTGAAGGTCGGAGTCA | 00000229239 | 199 bp |
| R | GACAAGCTTCCCGTTCTCAG | |||
| β-actin | F | TCTGGCACCACACCTTCTAC | 00000331789 | 169 bp |
| R | GATAGCACAGCCTGGATAGC | |||
| HPRT1 | F | CTGAGGATTTGGAAAGGGTG | 00000298556 | 156 bp |
| R | AATCCAGCAGGTCAGCAAAG | |||
| Β2M | F | CGCTACTCTCTCTTTCTGGC | 00000558401 | 133 bp |
| R | ATGTCGGATGGATGAAACCC | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Świerczewska, M.; Klejewski, A.; Brązert, M.; Kaźmierczak, D.; Iżycki, D.; Nowicki, M.; Zabel, M.; Januchowski, R. New and Old Genes Associated with Primary and Established Responses to Paclitaxel Treatment in Ovarian Cancer Cell Lines. Molecules 2018, 23, 891. https://doi.org/10.3390/molecules23040891
Świerczewska M, Klejewski A, Brązert M, Kaźmierczak D, Iżycki D, Nowicki M, Zabel M, Januchowski R. New and Old Genes Associated with Primary and Established Responses to Paclitaxel Treatment in Ovarian Cancer Cell Lines. Molecules. 2018; 23(4):891. https://doi.org/10.3390/molecules23040891
Chicago/Turabian StyleŚwierczewska, Monika, Andrzej Klejewski, Maciej Brązert, Dominika Kaźmierczak, Dariusz Iżycki, Michał Nowicki, Maciej Zabel, and Radosław Januchowski. 2018. "New and Old Genes Associated with Primary and Established Responses to Paclitaxel Treatment in Ovarian Cancer Cell Lines" Molecules 23, no. 4: 891. https://doi.org/10.3390/molecules23040891






