PER, a Circadian Clock Component, Mediates the Suppression of MMP-1 Expression in HaCaT Keratinocytes by cAMP
Abstract
:1. Introduction
2. Results
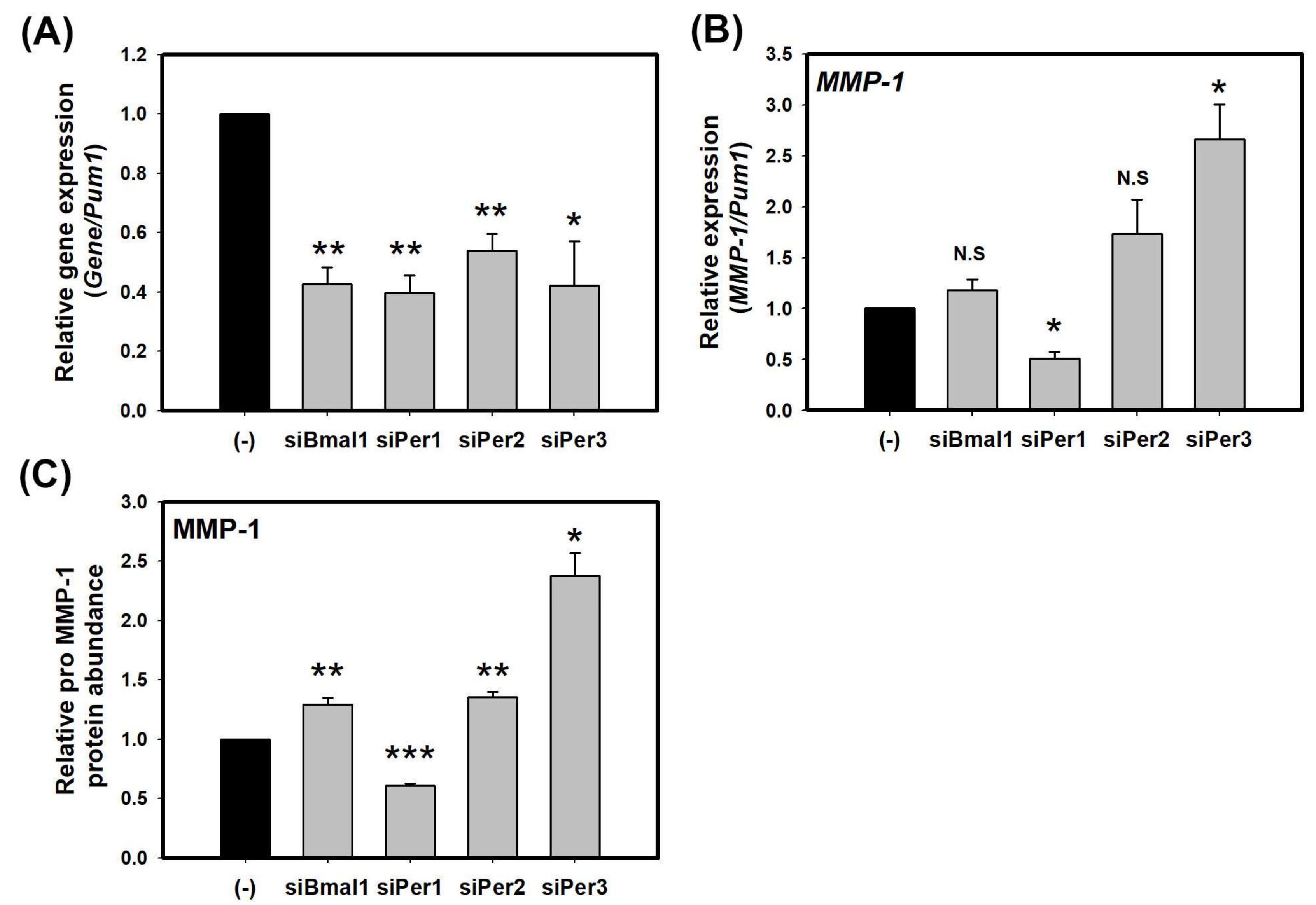
2.1. Knockdown of PER Proteins Increase the Expression of MMP-1 in HaCaT Cells
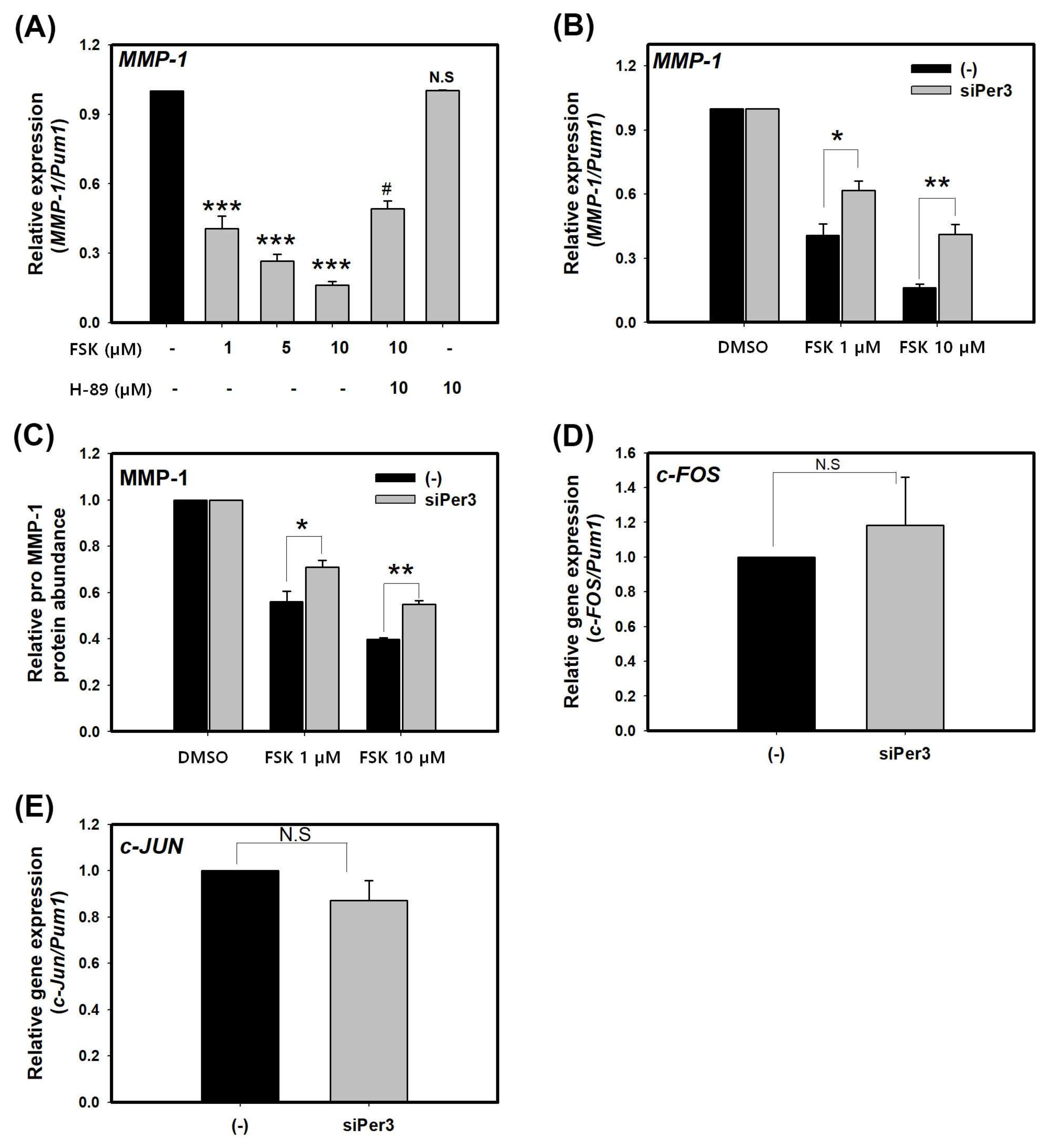
2.2. Suppression of MMP-1 by cAMP is Mediated by PER
2.3. LCE Increases PER Activity in HaCaT Cells
2.4. LCE Suppresses the Expression of MMP-1 through PER in HaCaT Cell
3. Discussion
4. Materials and Methods
4.1. Small-Interfering RNA Experiments and Pharmacological Treatment
4.2. RNA Isolation and Quantitative Real-Time RT–PCR
4.3. MMP1 Protein Determination by ELISA
4.4. Establishment of a Stable Cell Line for Per2/Bmal1 Promoter-Based Reporter Gene Assay
4.5. Bioluminescence Recording
4.6. Extraction of L. capitata
4.7. Pharmacological Treatment for Bioluminescence Recording
4.8. Determination of Cell Number
4.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef]
- Spoelstra, K.; Wikelski, M.; Daan, S.; Loudon, A.S.I.; Hau, M. Natural selection against a circadian clock gene mutation in mice. Proc. Natl. Acad. Sci. USA 2016, 113, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.; van Heerikhuize, J.J.; Wortel, J.; Buijs, R.M. A diurnal rhythm of stimulatory input to the hypothalamo-pituitary-adrenal system as revealed by timed intrahypothalamic administration of the vasopressin V1 antagonist. J. Neurosci. 1996, 16, 5555–5565. [Google Scholar] [PubMed]
- Ohdo, S.; Koyanagi, S.; Matsunaga, N.; Hamdan, A. Molecular basis of chronopharmaceutics. J. Pharm. Sci. 2011, 100, 3560–3576. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Marcacci, L.; Schibler, U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. CB 2000, 10, 1291–1294. [Google Scholar] [CrossRef]
- Pevet, P.; Challet, E. Melatonin: both master clock output and internal time-giver in the circadian clocks network. J. Physiol. Paris 2011, 105, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Storch, K.-F.; Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 15172–15177. [Google Scholar] [CrossRef] [PubMed]
- Schibler, U. Circadian time keeping: the daily ups and downs of genes, cells, and organisms. Prog. Brain Res. 2006, 153, 271–282. [Google Scholar] [PubMed]
- Brown, W.R. A review and mathematical analysis of circadian rhythms in cell proliferation in mouse, rat, and human epidermis. J. Investig. Dermatol. 1991, 97, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Henry, F.; Arrese, J.E.; Claessens, N.; Piérard-Franchimont, C.; Piérard, G.E. [Skin and its daily chronobiological clock]. Rev. Med. Liege 2002, 57, 661–665. [Google Scholar] [PubMed]
- Denda, M.; Tsuchiya, T. Barrier recovery rate varies time-dependently in human skin. Br. J. Dermatol. 2000, 142, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Le Fur, I.; Reinberg, A.; Lopez, S.; Morizot, F.; Mechkouri, M.; Tschachler, E. Analysis of circadian and ultradian rhythms of skin surface properties of face and forearm of healthy women. J. Investig. Dermatol. 2001, 117, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Yosipovitch, G.; Xiong, G.L.; Haus, E.; Sackett-Lundeen, L.; Ashkenazi, I.; Maibach, H.I. Time-dependent variations of the skin barrier function in humans: transepidermal water loss, stratum corneum hydration, skin surface pH, and skin temperature. J. Investig. Dermatol. 1998, 110, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Yosipovitch, G.; Sackett-Lundeen, L.; Goon, A.; Yiong Huak, C.; Leok Goh, C.; Haus, E. Circadian and ultradian (12 h) variations of skin blood flow and barrier function in non-irritated and irritated skin-effect of topical corticosteroids. J. Investig. Dermatol. 2004, 122, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Sandu, C.; Dumas, M.; Malan, A.; Sambakhe, D.; Marteau, C.; Nizard, C.; Schnebert, S.; Perrier, E.; Challet, E.; Pévet, P.; et al. Human skin keratinocytes, melanocytes, and fibroblasts contain distinct circadian clock machineries. Cell. Mol. Life Sci. CMLS 2012, 69, 3329–3339. [Google Scholar] [CrossRef] [PubMed]
- Grundschober, C.; Delaunay, F.; Pühlhofer, A.; Triqueneaux, G.; Laudet, V.; Bartfai, T.; Nef, P. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J. Biol. Chem. 2001, 276, 46751–46758. [Google Scholar] [CrossRef] [PubMed]
- Janich, P.; Toufighi, K.; Solanas, G.; Luis, N.M.; Minkwitz, S.; Serrano, L.; Lehner, B.; Benitah, S.A. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell 2013, 13, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, H.; Ramos, M.C.; Stuhlmann, D.; Sies, H.; Brenneisen, P. UVA-mediated downregulation of MMP-2 and MMP-9 in human epidermal keratinocytes. Biochem. Biophys. Res. Commun. 2003, 308, 486–491. [Google Scholar] [CrossRef]
- Vicentini, F.T.M.C.; He, T.; Shao, Y.; Fonseca, M.J.V.; Verri, W.A.; Fisher, G.J.; Xu, Y. Quercetin inhibits UV irradiation-induced inflammatory cytokine production in primary human keratinocytes by suppressing NF-κB pathway. J. Dermatol. Sci. 2011, 61, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Gaddameedhi, S.; Selby, C.P.; Kaufmann, W.K.; Smart, R.C.; Sancar, A. Control of skin cancer by the circadian rhythm. Proc. Natl. Acad. Sci. USA 2011, 108, 18790–18795. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.-H.; Reardon, J.T.; Kemp, M.; Sancar, A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA 2009, 106, 2864–2867. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.-H.; Reardon, J.T.; Sancar, A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011, 39, 3176–3187. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.R.; Fingleton, B.; Rothenberg, M.L.; Matrisian, L.M. Matrix metalloproteinases: biologic activity and clinical implications. J. Clin. Oncol. 2000, 18, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.; Qin, Z.; Xia, W.; Shao, Y.; Voorhees, J.J.; Fisher, G.J. Matrix-degrading Metalloproteinases in Photoaging. J. Investig. Dermatol. Symp. Proc. 2009, 14, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, C.-W.; Kim, E.K.; Lee, S.-J.; Park, N.-H.; Kim, H.-S.; Kim, H.-K.; Char, K.; Jang, Y.P.; Kim, J.-W. Inhibition effect of Gynura procumbens extract on UV-B-induced matrix-metalloproteinase expression in human dermal fibroblasts. J. Ethnopharmacol. 2011, 137, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Westermarck, J.; Li, S.; Jaakkola, P.; Kallunki, T.; Grénman, R.; Kähäri, V.-M. Activation of Fibroblast Collagenase-1 Expression by Tumor Cells of Squamous Cell Carcinomas Is Mediated by p38 Mitogen-activated Protein Kinase and c-Jun NH2-terminal Kinase-2. Cancer Res. 2000, 60, 7156–7162. [Google Scholar] [PubMed]
- Fagot, D.; Asselineau, D.; Bernerd, F. Direct role of human dermal fibroblasts and indirect participation of epidermal keratinocytes in MMP-1 production after UV-B irradiation. Arch. Dermatol. Res. 2002, 293, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.J.; Kang, S.; Varani, J.; Bata-Csorgo, Z.; Wan, Y.; Datta, S.; Voorhees, J.J. Mechanisms of photoaging and chronological skin aging. Arch. Dermatol. 2002, 138, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-H.; Moon, Y.; Shin, C.M.; Chung, J.H. Cyclic AMP suppresses matrix metalloproteinase-1 expression through inhibition of MAPK and GSK-3beta. J. Investig. Dermatol. 2010, 130, 2049–2056. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Li, A.; Hansen, K.F.; Cao, R.; Yoon, J.H.; Obrietan, K. CREB influences timing and entrainment of the SCN circadian clock. J. Biol. Rhythms 2010, 25, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, S.; Hamdan, A.M.; Horiguchi, M.; Kusunose, N.; Okamoto, A.; Matsunaga, N.; Ohdo, S. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 2011, 286, 32416–32423. [Google Scholar] [CrossRef] [PubMed]
- Obrietan, K.; Impey, S.; Smith, D.; Athos, J.; Storm, D.R. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J. Biol. Chem. 1999, 274, 17748–17756. [Google Scholar] [CrossRef] [PubMed]
- Tischkau, S.A.; Mitchell, J.W.; Tyan, S.-H.; Buchanan, G.F.; Gillette, M.U. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J. Biol. Chem. 2003, 278, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.M.; Stork, P.J.S. PKA phosphorylation of Src mediates cAMP’s inhibition of cell growth via Rap1. Mol. Cell 2002, 9, 85–94. [Google Scholar] [CrossRef]
- Mayr, B.M.; Canettieri, G.; Montminy, M.R. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl. Acad. Sci. USA 2001, 98, 10936–10941. [Google Scholar] [CrossRef] [PubMed]
- Houslay, M.D.; Kolch, W. Cell-type specific integration of cross-talk between extracellular signal-regulated kinase and cAMP signaling. Mol. Pharmacol. 2000, 58, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Okabe, T.; Kumagai, M.; Nakajima, Y.; Shirotake, S.; Kodaira, K.; Oyama, M.; Ueno, M.; Ikeda, M. The impact of HIF1α on the Per2 circadian rhythm in renal cancer cell lines. PLoS ONE 2014, 9, e109693. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, N.; Itcho, K.; Hamamura, K.; Ikeda, E.; Ikeyama, H.; Furuichi, Y.; Watanabe, M.; Koyanagi, S.; Ohdo, S. 24-hour rhythm of aquaporin-3 function in the epidermis is regulated by molecular clocks. J. Investig. Dermatol. 2014, 134, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, K.; Bae, I.-H.; Lee, S.H.; Jung, J.; Lee, T.R.; Cho, E.-G. TIMP3 is a CLOCK-dependent diurnal gene that inhibits the expression of UVB-induced inflammatory cytokines in human keratinocytes. FASEB J. 2017, 32, 1510–1523. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Ripperger, J.A.; Hoegger, D.C.; Bruegger, P.; Buch, T.; Birchler, T.; Mueller, A.; Albrecht, U.; Contaldo, C.; Brown, S.A. NONO couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. USA 2013, 110, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Kalfalah, F.; Janke, L.; Schiavi, A.; Tigges, J.; Ix, A.; Ventura, N.; Boege, F.; Reinke, H. Crosstalk of clock gene expression and autophagy in aging. Aging 2016, 8, 1876–1895. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-X.; Fu, X.-J.; Yang, K.; Chen, D.; Tang, H.; Zhao, Q.; Li, H.-X.; Fu, X.-J.; Yang, K.; Chen, D.; et al. The clock gene PER1 suppresses expression of tumor-related genes in human oral squamous cell carcinoma. Oncotarget 2016, 7, 20574–20583. [Google Scholar] [CrossRef] [PubMed]
- Climent, J.; Perez-Losada, J.; Quigley, D.A.; Kim, I.-J.; Delrosario, R.; Jen, K.-Y.; Bosch, A.; Lluch, A.; Mao, J.-H.; Balmain, A. Deletion of the PER3 Gene on Chromosome 1p36 in Recurrent ER-Positive Breast Cancer. J. Clin. Oncol. 2010, 28, 3770–3778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relógio, A.; Thomas, P.; Medina-Pérez, P.; Reischl, S.; Bervoets, S.; Gloc, E.; Riemer, P.; Mang-Fatehi, S.; Maier, B.; Schäfer, R.; et al. Ras-Mediated Deregulation of the Circadian Clock in Cancer. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Yang, N.; Borysiewicz, E.; Dudek, M.; Williams, J.L.; Li, J.; Maywood, E.S.; Adamson, A.; Hastings, M.H.; Bateman, J.F.; et al. Catabolic cytokines disrupt the circadian clock and the expression of clock-controlled genes in cartilage via an NFкB-dependent pathway. Osteoarthr. Cartil. 2015, 23, 1981–1988. [Google Scholar] [CrossRef] [PubMed]
- Kawara, S.; Mydlarski, R.; Shivji, G.; Tavadia, S.K.; Suzuki, H.; Mamelak, A.J.; Freed, I.; Wang, B.; Watanabe, H.; Bjarnason, G.A.; et al. Low-dose Ultraviolet B Rays Alter the mRNA Expression of the Circadian Clock Genes in Cultured Human Keratinocytes. J. Investig. Dermatol. 2002, 119, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Pagani, L.; Schmitt, K.; Meier, F.; Izakovic, J.; Roemer, K.; Viola, A.; Cajochen, C.; Wirz-Justice, A.; Brown, S.A.; Eckert, A. Serum factors in older individuals change cellular clock properties. Proc. Natl. Acad. Sci. USA 2011, 108, 7218–7223. [Google Scholar] [CrossRef] [PubMed]
- Antoch, M.P.; Kondratov, R.V. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb. Exp. Pharmacol. 2013, 217, 289–309. [Google Scholar]
- Banerjee, S.; Wang, Y.; Solt, L.A.; Griffett, K.; Kazantzis, M.; Amador, A.; El-Gendy, B.M.; Huitron-Resendiz, S.; Roberts, A.J.; Shin, Y.; et al. Pharmacological Targeting of the Mammalian Clock Regulates Sleep Architecture and Emotional Behavior. Nat. Commun. 2014, 5, 5759. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, P.S.; Ahern, S.A.; Smith, L.C.; da Silva Santos, C.S.; Wager, T.T.; Bechtold, D.A. Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, A.; Misawa, K.; Ikeda, Y.; Haraguchi, A.; Kamagata, M.; Tahara, Y.; Shibata, S. Potent Effects of Flavonoid Nobiletin on Amplitude, Period, and Phase of the Circadian Clock Rhythm in PER2::LUCIFERASE Mouse Embryonic Fibroblasts. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Spörl, F.; Schellenberg, K.; Blatt, T.; Wenck, H.; Wittern, K.-P.; Schrader, A.; Kramer, A. A Circadian Clock in HaCaT Keratinocytes. J. Investig. Dermatol. 2011, 131, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Ongena, K.; Das, C.; Smith, J.L.; Gil, S.; Johnston, G. Determining cell number during cell culture using the Scepter cell counter. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.-J.; Wang, W.-C.; Wang, C.-K.; Wu, C.-H.; Yang, M.-D.; Chang, Y.-J.; Jian, J.-Y.; Tai, C.-J. Fermented Wheat Germ Extract Induced Cell Death and Enhanced Cytotoxicity of Cisplatin and 5-Fluorouracil on Human Hepatocellular Carcinoma Cells. Evid.-Based Complement. Altern. Med. ECAM 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds Lespedeza capitate extract are available from the authors. |
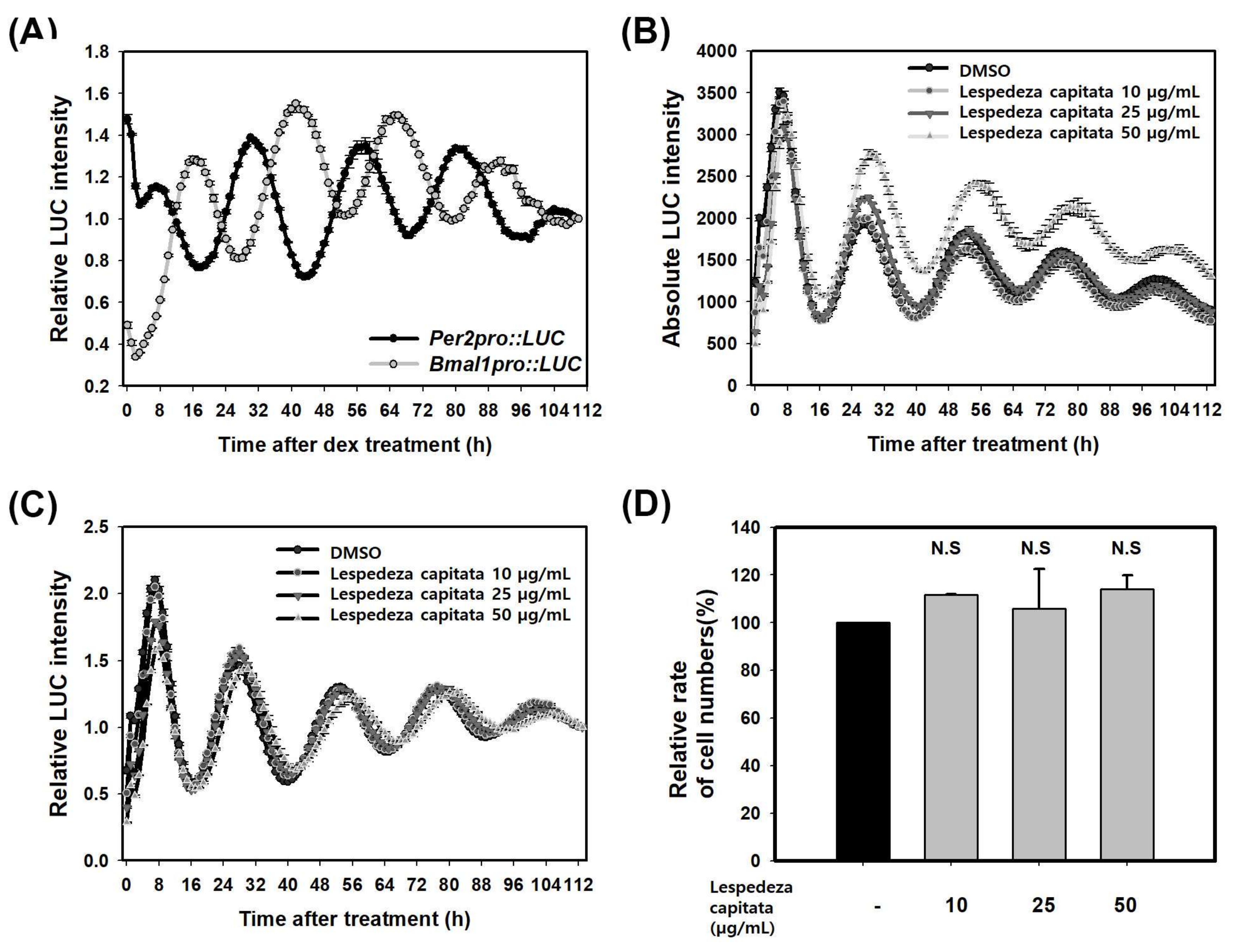

| Per2pro::LUC | Bmal1pro::LUC | ||||
| Period (h) | Average | 25.48 | 25.36 | ||
| S.E | 0.17 | 0.07 | |||
| Amplitude | Average | 0.24 | 0.26 | ||
| S.E | 0.01 | 0.00 | |||
| R.A.E | Average | 0.12 | 0.13 | ||
| S.E | 0.00 | 0.01 | |||
| Phase (h) | Average | 7.25 | 16.50 | ||
| S.E | 0.25 | 0.29 | |||
| DMSO | Lespedeza Capitata | ||||
| 10 µg/mL | 25 µg/mL | 50 µg/mL | |||
| Period (h) | Average | 24.87 | 24.72 | 25.01 | 25.53 |
| S.E | 0.02 | 0.03 | 0.01 | 0.04 | |
| Amplitude | Average | 0.29 | 0.28 | 0.27 | 0.22 |
| S.E | 0.00 | 0.00 | 0.00 | 0.00 | |
| R.A.E. | Average | 0.16 | 0.17 | 0.17 | 0.19 |
| S.E | 0.00 | 0.00 | 0.00 | 0.00 | |
| Phase (h) | Average | 7 | 7 | 7 | 8 |
| S.E | 0 | 0 | 0 | 0 | |
| Gene Name | Accession Number | Primer Sequence | |
|---|---|---|---|
| PUM1 | NM_014676 | hPum1-F | 5′-CGGTCGTCCTGAGGATAAAA-3′ |
| hPum1-R | 5′-CGTACGTGAGGCGTGAGTAA-3′ | ||
| BMAL1 | NM_001668 | hBmal1-F | 5′-TTAAGAGGTGCCACCAATCC-3′ |
| hBmal1-R | 5′-CTTCCCTCGGTCACATCCTA-3′ | ||
| PER1 | NM_002616 | hPer1-F | 5′-GGACACTCCTGCGACCAG-3′ |
| hPer1-R | 5′-GGGAGTGAGGTGGAAGATCTAA-3′ | ||
| PER2 | NM_022817.2 | hPer2-F | 5′-TTCCCAGCAAACGTCCCAG-3′ |
| hPer2-R | 5′-GGTGCGTACCTACTCCCGT-3′ | ||
| PER3 | NM_016831 | hPer3-F | 5′-GCGCATTCTCATGACATACC-3′ |
| hPer3-R | 5′-TGCTGCTGCCTCATACTTTC-3′ | ||
| c-FOS | NM_005252.3 | hc-Fos-F | 5′-CTACCACTCACCCGCAGACT-3′ |
| hc-Fos-R | 5′-AGGTCCGTGCAGAAGTCCT-3′ | ||
| c-JUN | NM_002228.3 | hc-Jun-F | 5′-CCAAAGGATAGTGCGATGTTT-3′ |
| hc-Jun-R | 5′-CTGTCCCTCTCCACTGCAAC-3′ | ||
| MMP-1 | NM_002421.3 | Qiagen (PPH00120B) | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeom, M.; Lee, H.; Shin, S.; Park, D.; Jung, E. PER, a Circadian Clock Component, Mediates the Suppression of MMP-1 Expression in HaCaT Keratinocytes by cAMP. Molecules 2018, 23, 745. https://doi.org/10.3390/molecules23040745
Yeom M, Lee H, Shin S, Park D, Jung E. PER, a Circadian Clock Component, Mediates the Suppression of MMP-1 Expression in HaCaT Keratinocytes by cAMP. Molecules. 2018; 23(4):745. https://doi.org/10.3390/molecules23040745
Chicago/Turabian StyleYeom, Miji, HansongI Lee, Seoungwoo Shin, Deokhoon Park, and Eunsun Jung. 2018. "PER, a Circadian Clock Component, Mediates the Suppression of MMP-1 Expression in HaCaT Keratinocytes by cAMP" Molecules 23, no. 4: 745. https://doi.org/10.3390/molecules23040745





