Analysis of Protein-Phenolic Compound Modifications Using Electrochemistry Coupled to Mass Spectrometry
Abstract
:1. Introduction
2. Results
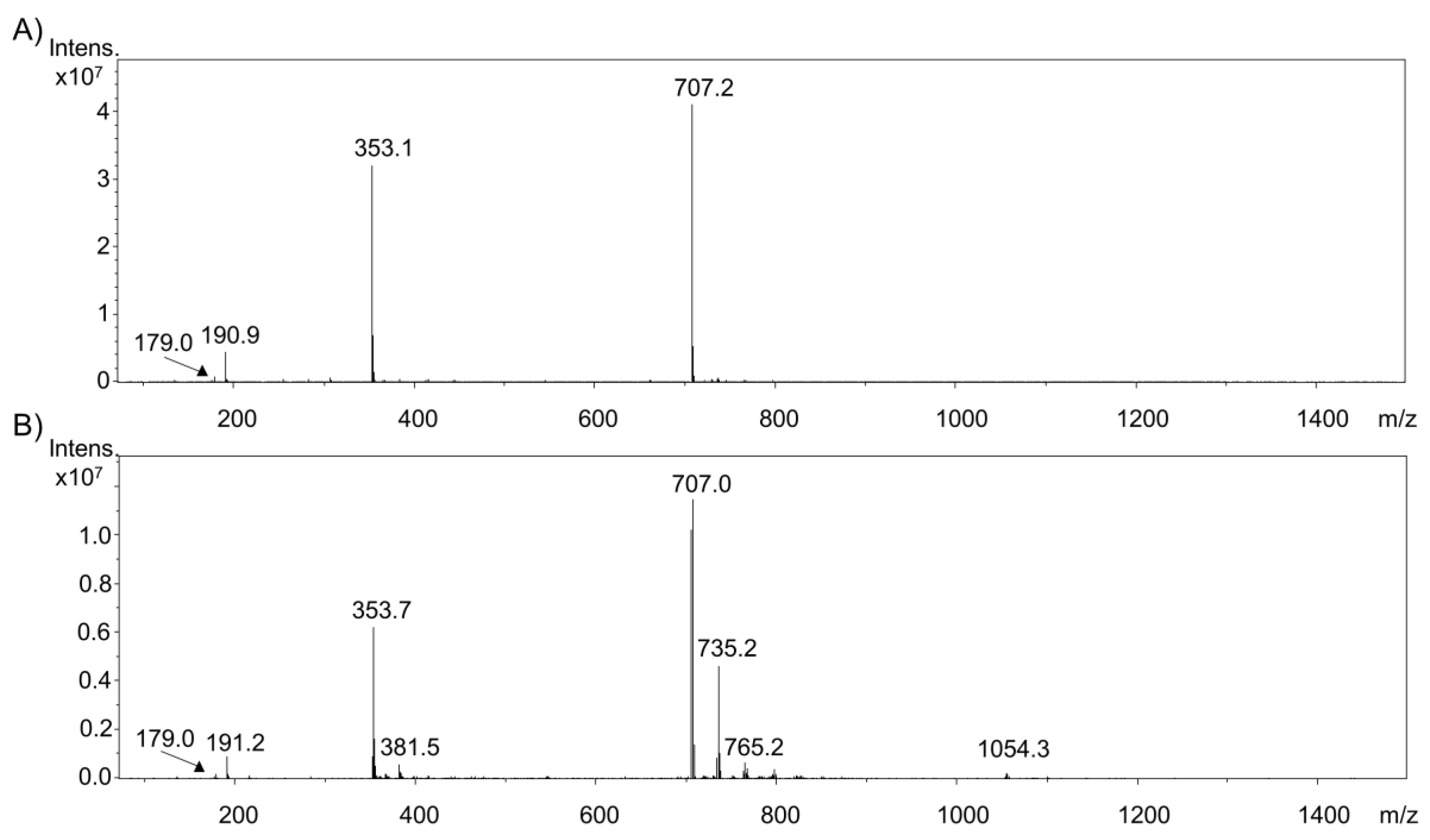
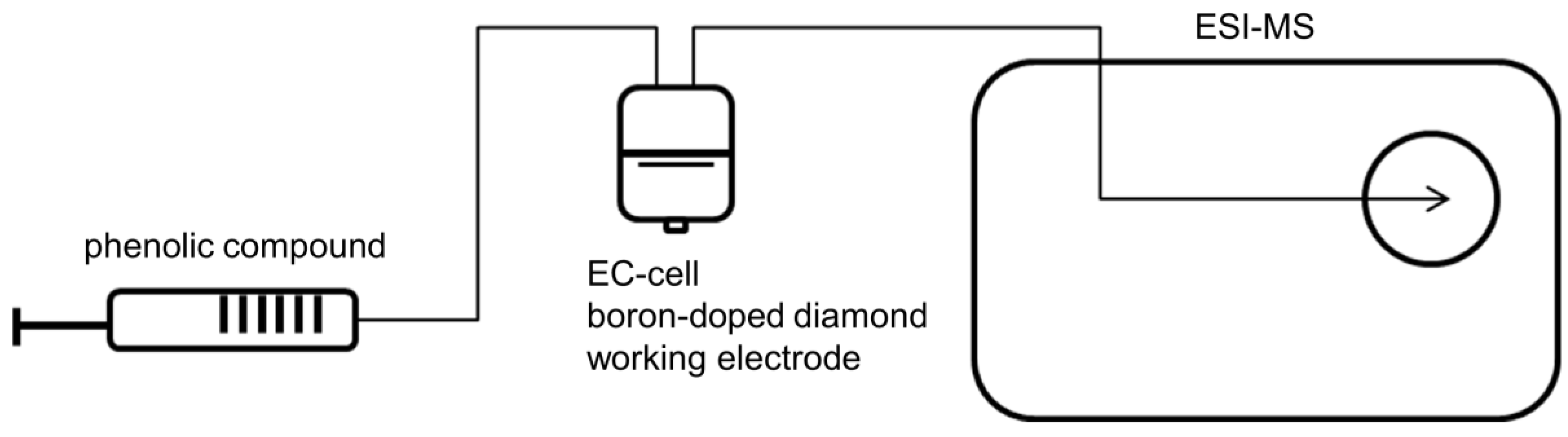
2.1. Electrochemical Oxidation of Phenolic Acids by Means of EC/ESI-MS
2.2. Comparison of the Oxidation of Various Phenolic Acids
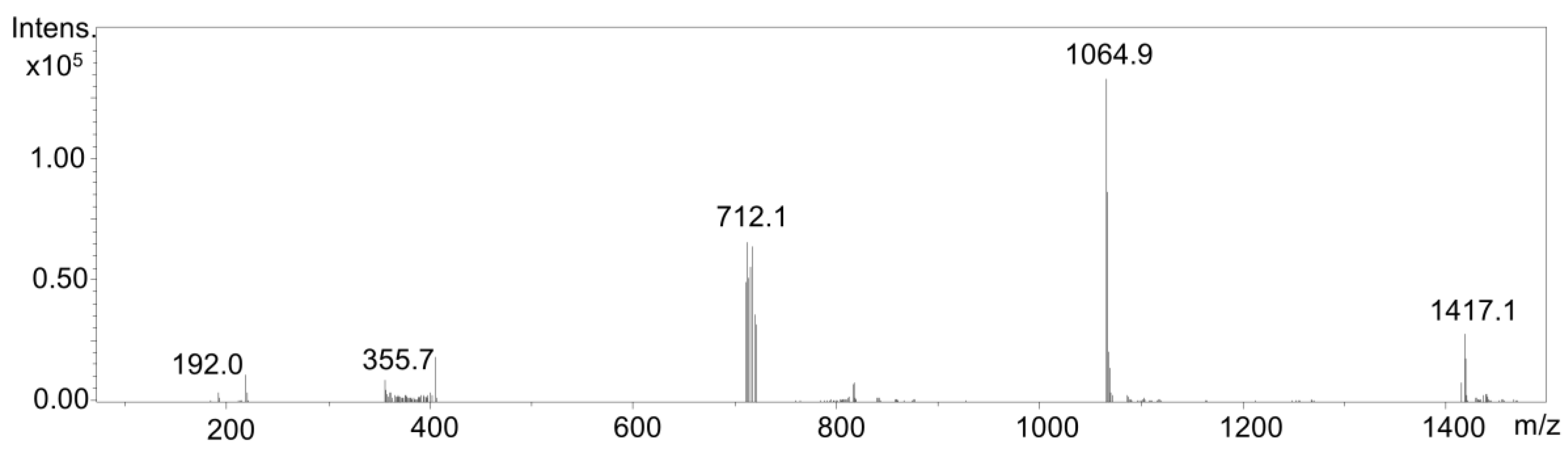
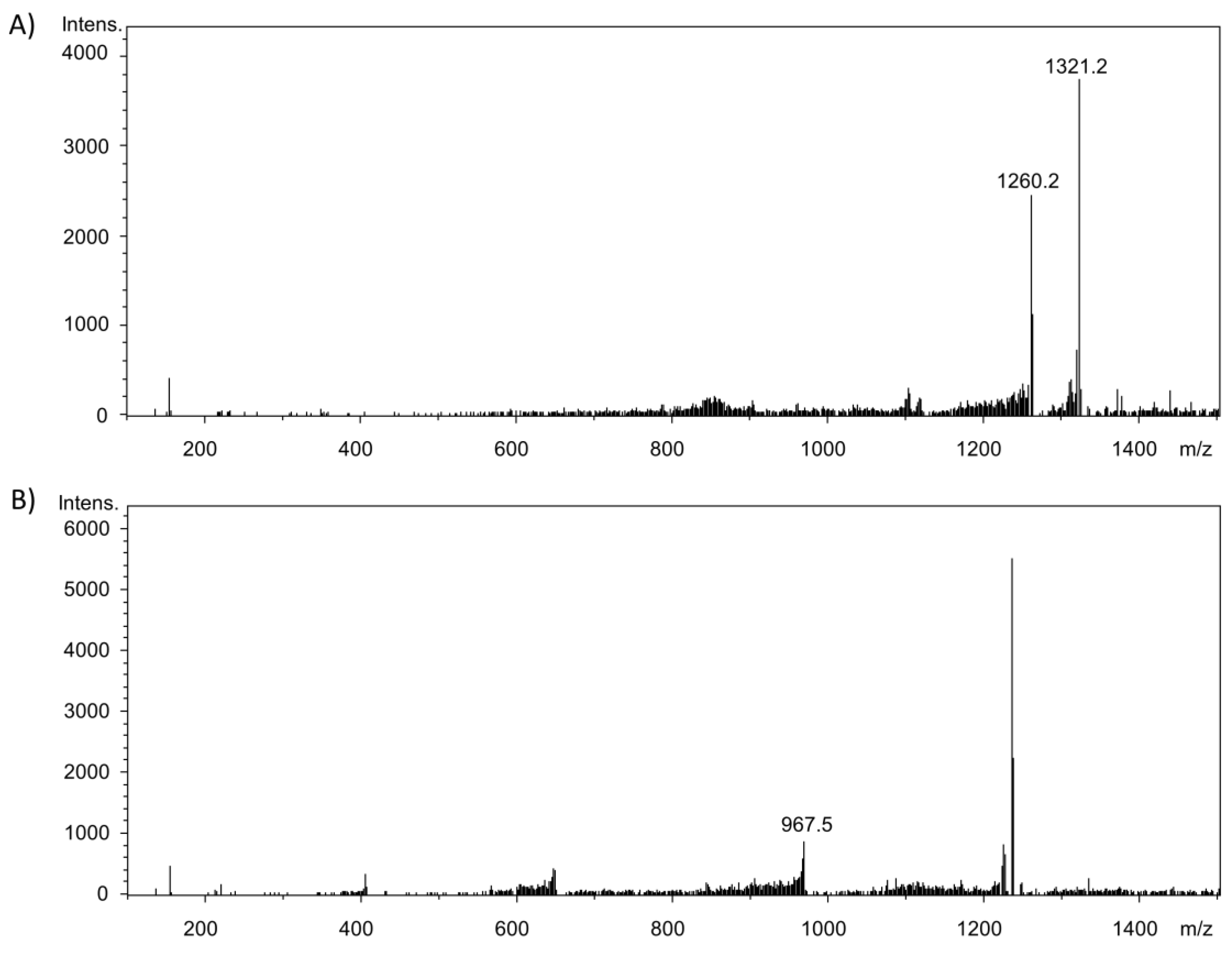
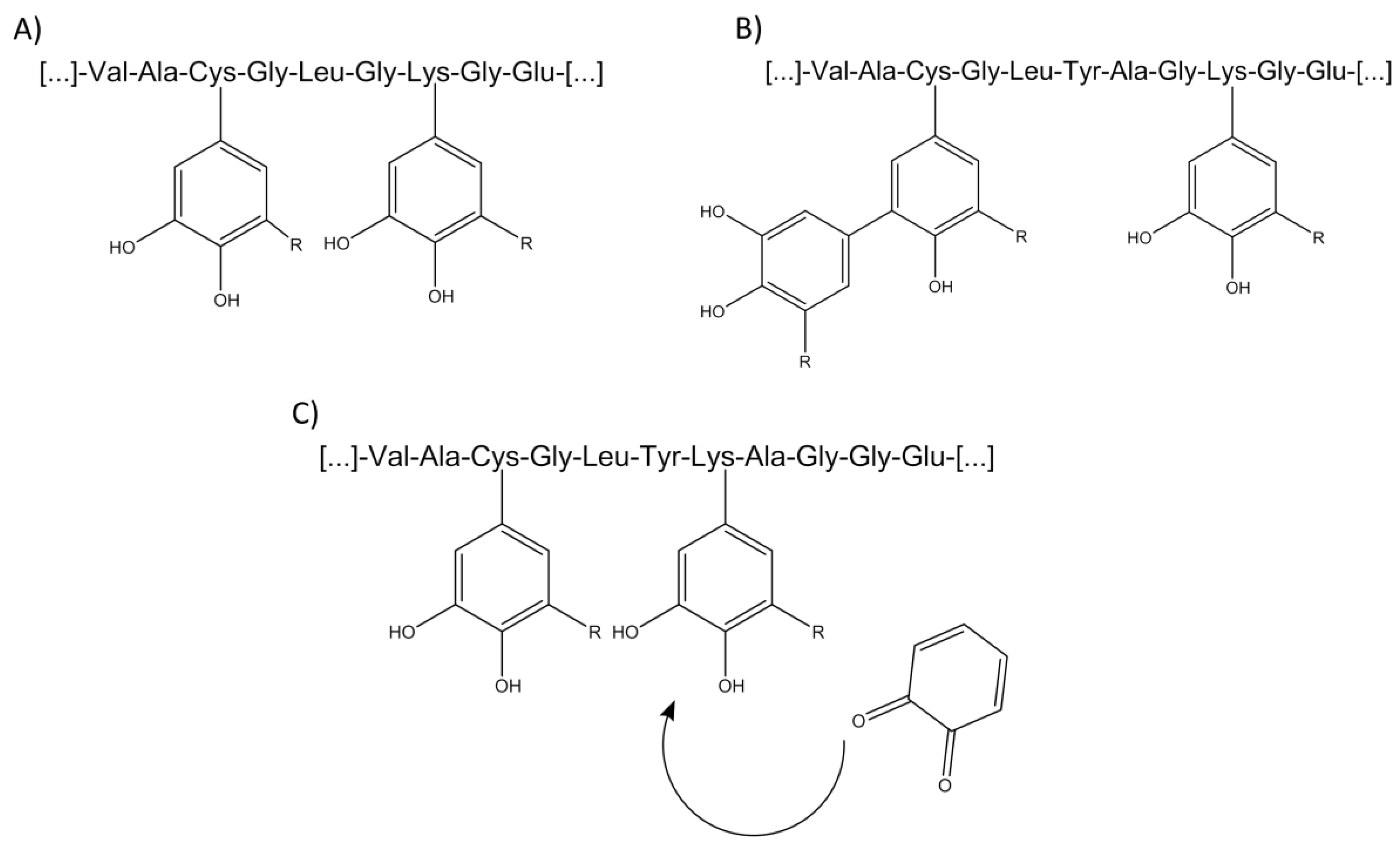
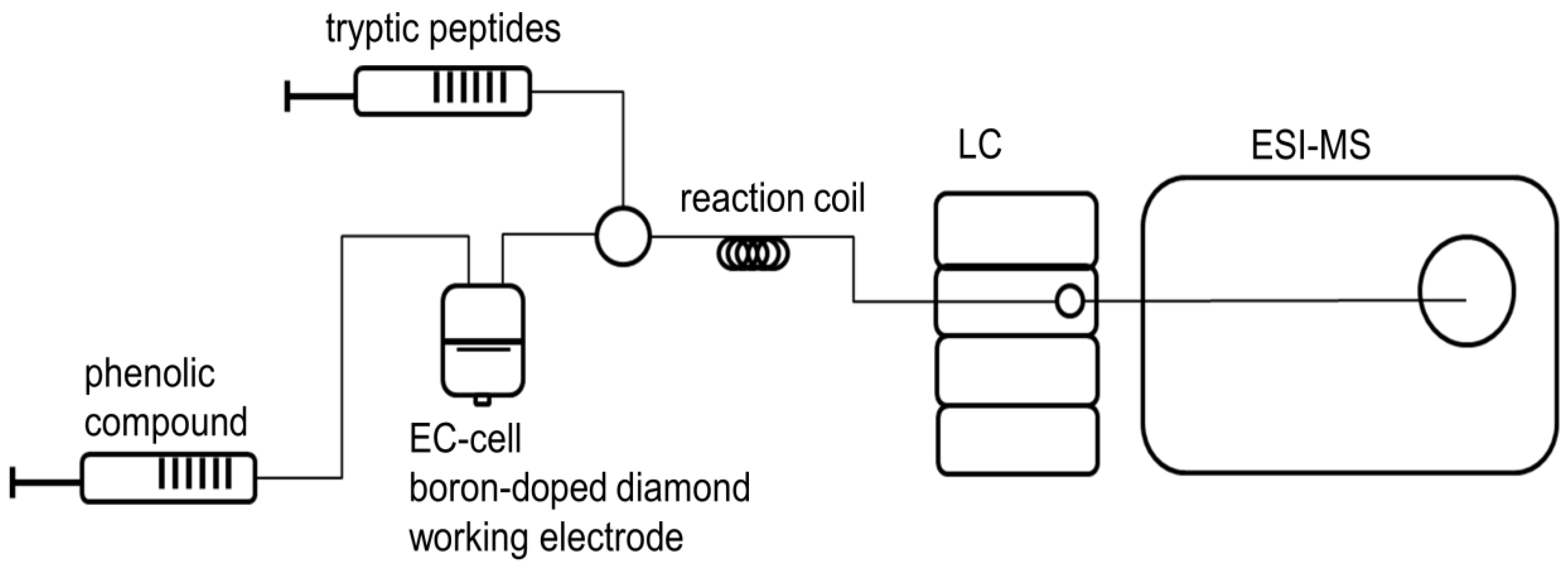
2.3. Investigation of Adduct Formation of Chlorogenic Acid and Alpha-Lactalbumin Using EC/LC/ESI-MS
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Electrochemical Oxidation of Phenolic Acids by Means of EC/MS
4.3 Tryptic Digestion of Alpha-Lactalbumin
4.4 Investigation of Adduct Formation of Chlorogenic Acid and Alpha-Lactalbumin Using EC/LC/ESI-MS
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Le Bourvellec, C.; Renard, C. Interactions between polyphenols and macromolecules: Quantification methods and mechanisms. Crit. Rev. Food Sci. Nutr. 2012, 52, 213–248. [Google Scholar] [CrossRef] [PubMed]
- Rohn, S.; Rawel, H.M.; Kroll, J. Inhibitory effects of plant phenols on the activity of selected enzymes. J. Agric. Food Chem. 2002, 50, 3566–3571. [Google Scholar] [CrossRef]
- Petzke, K.J.; Schuppe, S.; Rohn, S.; Rawel, H.M.; Kroll, J. Chlorogenic acid moderately decreases the quality of whey proteins in rats. J. Agric. Food Chem. 2005, 53, 3714–3720. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.J.; Smales, C.M.; James, D.C. How can thermal processing modify the antigenicity of proteins? Allergy 2001, 56, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Cilliers, J.J.; Singleton, V.L. Characterization of the products of nonenzymic autoxidative phenolic reactions in a caffeic acid model system. J. Agric. Food Chem. 1991, 39, 1298–1303. [Google Scholar] [CrossRef]
- Buchner, N.; Krumbein, A.; Rohn, S.; Kroh, L.W. Effect of thermal processing on the flavonols rutin and quercetin. Rapid Commun. Mass Spectrom. 2006, 20, 3229–3235. [Google Scholar] [CrossRef] [PubMed]
- Thurston, C.F. The structure and function of fungal laccases. Microbiology 1994, 140, 19–26. [Google Scholar] [CrossRef]
- Namiki, M.; Yabuta, G.; Koizumi, Y.; YANo, M. Development of free radical products during the greening reaction of caffeic acid esters (or chlorogenic acid) and a primary amino compound. Biosci. Biotechnol. Biochem. 2001, 65, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Kroll, J.; Rawel, H.M.; Seidelmann, N. Physicochemical properties and susceptibility to proteolytic digestion of myoglobin-phenol derivatives. J. Agric. Food Chem. 2000, 48, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Kai, G. A review of dietary polyphenol-plasma protein interactions: Characterization, influence on the bioactivity, and structure-affinity relationship. Crit. Rev. Food Sci. Nutr. 2012, 52, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Rohn, S. Possibilities and limitations in the analysis of covalent interactions between phenolic compounds and proteins. Food Res. Int. 2014, 65, 13–19. [Google Scholar] [CrossRef]
- Büter, L.; Frensemeier, L.M.; Vogel, M.; Karst, U. Dual reductive/oxidative electrochemistry/liquid chromatography/mass spectrometry: Towards peptide and protein modification, separation and identification. J. Chromatogr. A 2017, 1479, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Jahn, S.; Faber, H.; Zazzeroni, R.; Karst, U. Electrochemistry/liquid chromatography/mass spectrometry to demonstrate irreversible binding of the skin allergen p-phenylenediamine to proteins. Rapid Commun. Mass Spectrom. 2012, 26, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, R.; Yamaguchi, M.; Hotta, H.; Osakai, T.; Kimoto, T. Product analysis of caffeic acid oxidation by on-line electrochemistry/electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004, 15, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Saint-Cricq de Gaulejac, N.; Provost, C.; Vivas, N. Comparative study of polyphenol scavenging activities assessed by different methods. J. Agric. Food Chem. 1999, 47, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Wal, J.M. Cow’s milk proteins/allergens. Ann. Allergy Asthma Immunol. 2002, 89, 3–10. [Google Scholar] [CrossRef]
- Hendrix, T.M.; Griko, Y.; Privalov, P. Energetics of structural domains in α-lactalbumin. Protein Sci. 1996, 5, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Büter, L.; Faber, H.; Wigger, T.; Vogel, M.; Karst, U. Differential protein labeling based on electrochemically generated reactive intermediates. Anal. Chem. 2015, 87, 9931–9938. [Google Scholar] [CrossRef] [PubMed]
- Cilliers, J.J.; Singleton, V.L. Nonenzymic autoxidative phenolic browning reactions in a caffeic acid model system. J. Agric. Food Chem. 1989, 37, 890–896. [Google Scholar] [CrossRef]
- Büter, L.; Vogel, M.; Karst, U. Adduct formation of electrochemically generated reactive intermediates with biomolecules. TrAC 2015, 70, 74–91. [Google Scholar] [CrossRef]
- Olthof, M.R.; Hollman, P.C.; Katan, M.B. Chlorogenic acid and caffeic acid are absorbed in humans. J. Nutr. 2001, 131, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Prigent, S.V.; Gruppen, H.; Visser, A.J.; Van Koningsveld, G.A.; De Jong, G.A.; Voragen, A.G. Effects of non-covalent interactions with 5-O-caffeoylquinic acid (chlorogenic acid) on the heat denaturation and solubility of globular proteins. J. Agric. Food Chem. 2003, 51, 5088–5095. [Google Scholar] [CrossRef] [PubMed]
- Glabasnia, A.; Hofmann, T. Sensory-directed identification of taste-active ellagitannins in American (Quercus alba L.) and European oak wood (Quercus robur L.) and quantitative analysis in bourbon whiskey and oak-matured red wines. J. Agric. Food Chem. 2006, 9, 3380–3390. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.; Meisel, H.; Creminon, C.; Wal, J. Post-translational phosphorylation affects the IgE binding capacity of caseins. FEBS Lett. 2000, 467, 239–244. [Google Scholar] [CrossRef]
- Brudzynski, K.; Sjaarda, C.; Maldonado-Alvarez, L. A new look on protein-polyphenol complexation during honey storage: Is this a random or organized event with the help of dirigent-like proteins? PLoS ONE 2013, 8, e72897. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, B.; Premovic, P.; Sealy, R.C. Semiquinone anion radicals from addition of amino acids, peptides, and proteins to quinones derived from oxidation of catechols and catecholamines. An ESR spin stabilization study. J. Biol. Chem. 1987, 262, 11080–11087. [Google Scholar] [PubMed]
- Takizawa, Y.; Munakata, T.; Iwasa, Y.; Suzuki, T.; Mitsuhashi, T. Novel oxidative coupling of monophenols in the system of cupric chloride-oxygen-alcohol. J. Org. Chem. 1985, 50, 4383–4386. [Google Scholar] [CrossRef]
- Wang, T.; Chen, J.H.; Hsiang, W.M. Catalytic systhesis of humic acids containing various amino acids and dipeptides. Soil Sci. 1985, 140, 3–10. [Google Scholar] [CrossRef]
- Morschheuser, L.; Mink, K.; Horst, R.; Kallinich, C.; Rohn, S. Immunological analysis of food proteins using High-performance thin-layer chromatography-immunostaining. J. Chromatogr. A 2017, 1526, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Morschheuser, L.; Wessels, H.; Pille, C.; Fischer, J.; Hünniger, T.; Fischer, M.; Paschke-Kratzin, A.; Rohn, S. HPTLC-aptastaining–Innovative protein detection system for high-performance thin-layer chromatography. Sci. Rep. 2016, 6, 26665. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available. |
| Fragments [m/z] | Position | Sequence |
|---|---|---|
| 1198.6 | 118-127 | Val-Gly-Ile-Asn-Tyr-Trp-Leu-Ala-His-Lys |
| 749.4 | 113-118 | Glu-Leu-Lys-Asp-Leu-Lys |
| 652.3 | 25-29 | Cys-Glu-Val-Phe-Arg |
| 616.3 | 20-24 | Glu-Gln-Leu-Thr-Lys |
| 544.3 | 78-81 | Ile-Trp-Cys-Lys |
| 486.3 | 114-117 | Ile-Leu-Asp-Lys |
| 387.2 | 30-32 | Glu-Leu-Lys |
| 373.2 | 33-35 | Asp-Leu-Lys |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kallinich, C.; Schefer, S.; Rohn, S. Analysis of Protein-Phenolic Compound Modifications Using Electrochemistry Coupled to Mass Spectrometry. Molecules 2018, 23, 264. https://doi.org/10.3390/molecules23020264
Kallinich C, Schefer S, Rohn S. Analysis of Protein-Phenolic Compound Modifications Using Electrochemistry Coupled to Mass Spectrometry. Molecules. 2018; 23(2):264. https://doi.org/10.3390/molecules23020264
Chicago/Turabian StyleKallinich, Constanze, Simone Schefer, and Sascha Rohn. 2018. "Analysis of Protein-Phenolic Compound Modifications Using Electrochemistry Coupled to Mass Spectrometry" Molecules 23, no. 2: 264. https://doi.org/10.3390/molecules23020264




