Jasmonate-Elicited Stress Induces Metabolic Change in the Leaves of Leucaena leucocephala
Abstract
:1. Introduction
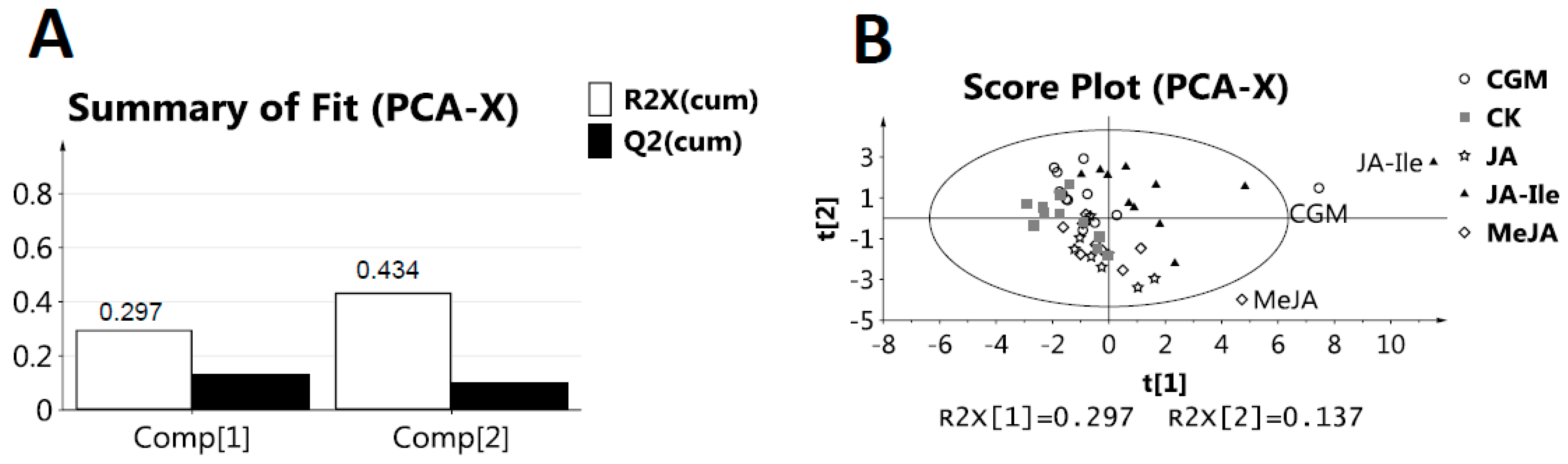
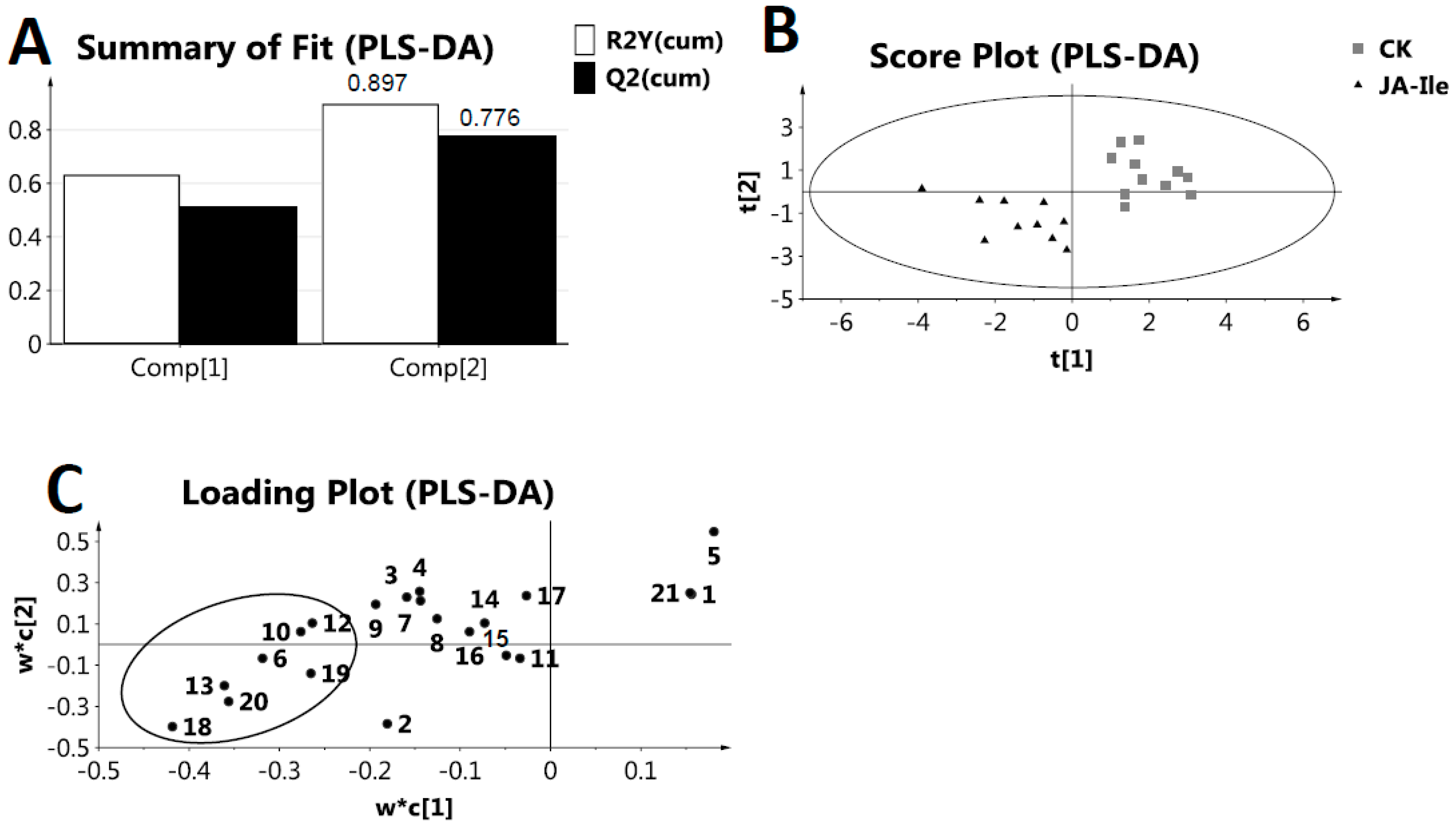
2. Results and Discussion
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Growth, Treatment and Harvest
3.3. Sample Preparation
3.4. NMR Experiments
3.5. NMR Data Processing and Multivariate Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| JA | jasmonic acid |
| MeJA | methyl jasmonate |
| CGM | 2-[(6-ethyl-1-oxoindane-4-carbonyl) amino] acetic acid methyl ester |
| JA-Ile | jasmonoyl-l-isoleucine |
| NMR | nuclear magnetic resonance |
| PCA | principal component analysis |
| PLS-DA | partial least squares discriminant analysis |
| TSP | sodium salt of 3-trimethylsilylpropionic acid |
References
- Negi, V.S.; Pal, A.; Singh, R.; Borthakur, D. Identification of species-specific genes from Leucaena leucocephala using interspecies suppression subtractive hybridisation. Ann. Appl. Biol. 2011, 159, 387–398. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Jones, J.D.G. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Abel, C.; Sohrabi, R.; Petri, J.; Haupt, I.; Cosimano, J.; Gershenzon, J.; Tholl, D. Variation of herbivore-induced volatile terpenes among arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, tps02 and tps03. Plant Physiol. 2010, 153, 1293–1310. [Google Scholar] [CrossRef] [PubMed]
- Schuler, G.; Mithofer, A.; Baldwin, I.T.; Berger, S.; Ebel, J.; Santos, J.G.; Herrmann, G.; Holscher, D.; Kramell, R.; Kutchan, T.M.; et al. Coronalon: A powerful tool in plant stress physiology. Febs. Lett. 2004, 563, 17–22. [Google Scholar] [CrossRef]
- Berim, A.; Spring, O.; Conrad, J.; Maitrejean, M.; Boland, W.; Petersen, M. Enhancement of lignan biosynthesis in suspension cultures of linum nodiflorum by coronalon, indanoyl-isoleucine and methyl jasmonate. Planta 2005, 222, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.W.; Xu, Q.L.; Wang, J.; Ren, H.; Zheng, M.F. Preparation of 2-[(6-ethyl-1-oxoindane-4-carbonyl)amino]acetic Acid Methyl Ester as Plant Defense Inducer. U.S. Patent CN 103113255 A, 22 May 2013. [Google Scholar]
- Hamberg, M.; Gardner, H.W. Oxylipin pathway to jasmonates-biochemistry and biological significance. Biochim. Et. Biophys. Acta 1992, 1165, 1–18. [Google Scholar] [CrossRef]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Robert, C.A.M.; Turlings, T.C.J.; Rudaz, S.; Wolfender, J.L. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Chen, F.; Wang, X.; Rajapakse, N.C. Effect of methyl jasmonate on secondary metabolites of sweet basil (Ocimum basilicum L.). J. Agric. Food Chem. 2006, 54, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Aerts, R.J.; Gisi, D.; Decarolis, E.; Deluca, V.; Baumann, T.W. Methyl jasmonate vapor incerease the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J. 1994, 5, 635–643. [Google Scholar] [CrossRef]
- Wei, S. Methyl jasmonic acid induced expression pattern of terpenoid indole alkaloid pathway genes in Catharanthus Roseus seedlings. Plant Growth Regul. 2010, 61, 243–251. [Google Scholar] [CrossRef]
- Chen, H.; Jones, A.D.; Howe, G.A. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett. 2006, 580, 2540–2546. [Google Scholar] [CrossRef]
- Vestena, S.; Fett-Neto, A.G.; Duarte, R.C.; Ferreira, A.G. Regulation of mimosine accumulation in Leucaena Leucocephala seedlings. Plant Sci. 2001, 161, 597–604. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. Biomagresbank. Nucleic Acids. Res. 2008, 36 (Suppl. S1), D402–D408. [Google Scholar] [CrossRef]
- Nokihara, K.; Hirata, A.; Sogon, T.; Ohyama, T. Preparative scale isolation, purification and derivatization of mimosine, a non-proteinogenic amino acid. Amino Acids 2012, 43, 475–482. [Google Scholar] [CrossRef]
- Tao, L.; Huang, J.; Zhao, Y.; Li, C. Chemical constituents in Buddleja albiflora. Zhongguo Zhongyao Zazhi 2009, 34, 3043–3046. [Google Scholar]
- SORO, Y.; KASSI, A.B.B.; BAMBA, F.; SIAKA, S.; Touré, S.A.; COUSTARD, J.-M. Flavonoids and gallic acid from leaves of Santaloides afzelii (connaraceae). Rasayan J. Chem. 2012, 5, 332–337. [Google Scholar]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Staswick, P.E. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in arabidopsis. Plant Cell Online 2004, 16, 2117–2127. [Google Scholar] [CrossRef]
- Tamogami, S.; Rakwal, R.; Agrawal, G.K. Interplant communication: Airborne methyl jasmonate is essentially converted into ja and ja-ile activating jasmonate signaling pathway and vocs emission. Biochem. Biophys. Res. Commun. 2008, 376, 723–727. [Google Scholar] [CrossRef]
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-l-isoleucine by multiple members of the cytochrome p450 94 family in arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738. [Google Scholar] [CrossRef] [PubMed]
- Elkahoui, S.; Smaoui, A.; Zarrouk, M.; Ghrir, R.; Limam, H. Salt-induced lipid changes in Catharanthus roseus cultured cell suspensions. Phytochemistry 2004, 65, 1911–1917. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.C.Q.; Tawata, S. The chemistry and biological activities of mimosine: A review. Phytother. Res. 2016, 30, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Xuan, T.D.; Elzaawely, A.A.; Deba, F.; Fukuta, M.; Tawata, S. Mimosine in leucaena as a potent bio-herbicide. Agrono. Sustain. Dev. 2006, 26, 89–97. [Google Scholar] [CrossRef]
- Allison, M.J.; Hammond, A.C.; Jones, R.J. Detection of ruminal bacteria that degrade toxic dihydroxypyridine compounds produced from mimosine. Appl. Environ. Microbiol. 1990, 56, 590–594. [Google Scholar] [PubMed]
- Hammond, A.C. Leucaena toxicosis and its control in ruminants. J. Anim. Sci. 1995, 73, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Kramell, R.; Schmidt, J.; Schneider, G.; Sembdner, G.; Schreiber, K. Synthesis of N-(jasmonoyl) amino acid conjugates. Tetrahedron 1988, 44, 5791–5807. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.G.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for nmr spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.M.; Klein, M.S.; Hochrein, J.; Oefner, P.J.; Spang, R.; Gronwald, W. State of the art data normalization methods improve nmr-based metabolomic analysis. Metabolomics 2012, 8, S146–S160. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 14–16 are available from the authors. |
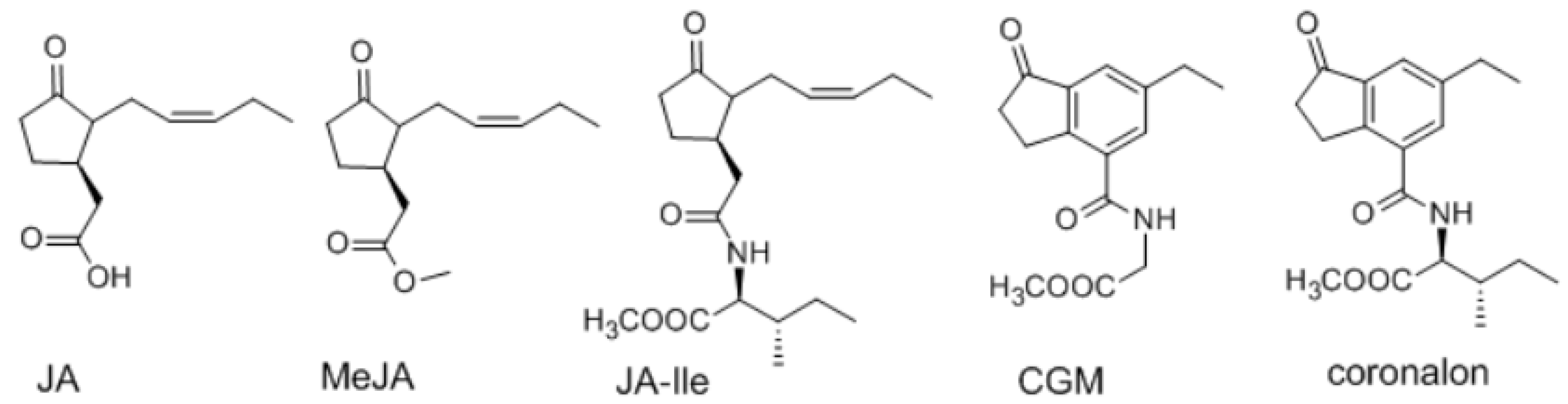
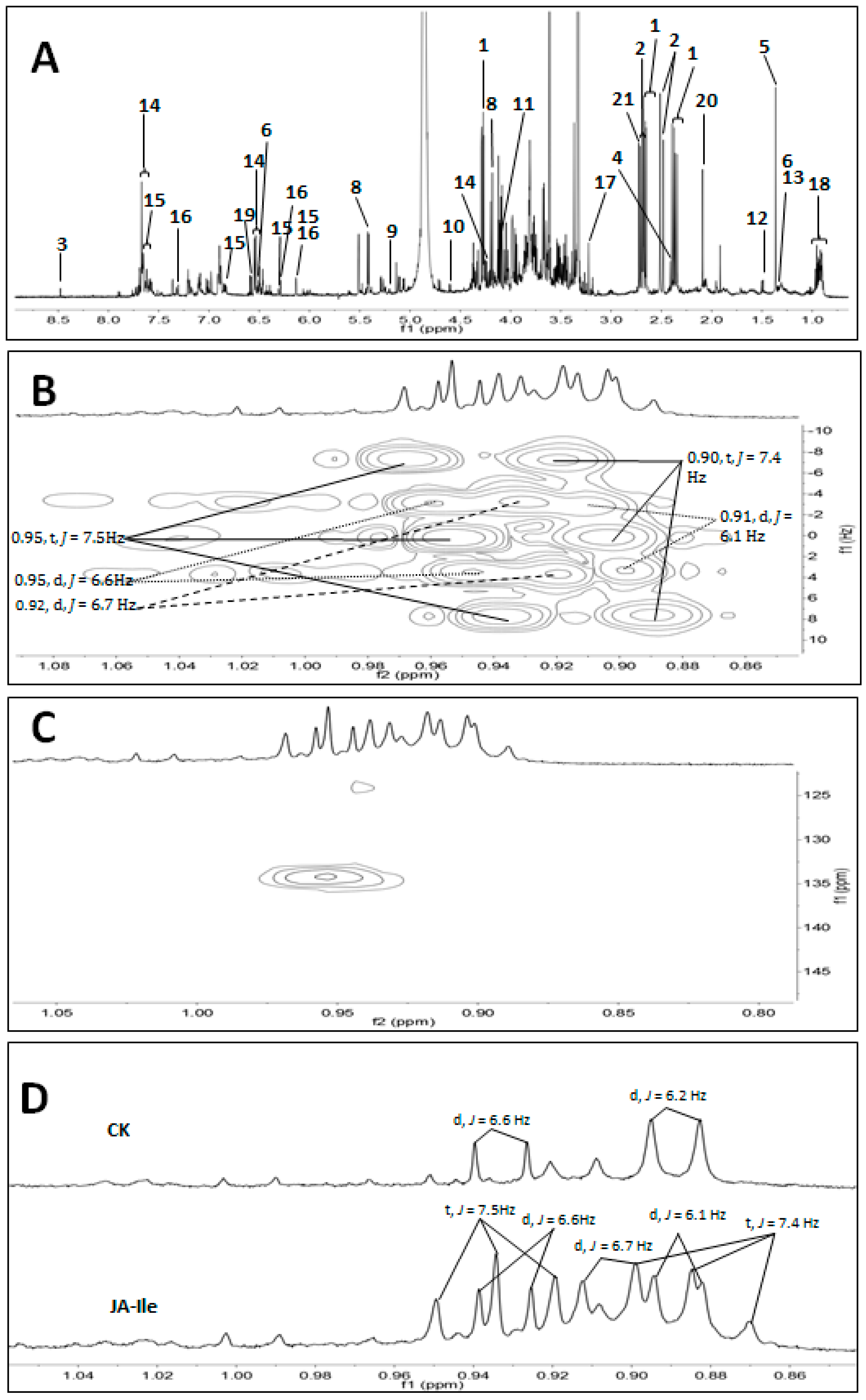
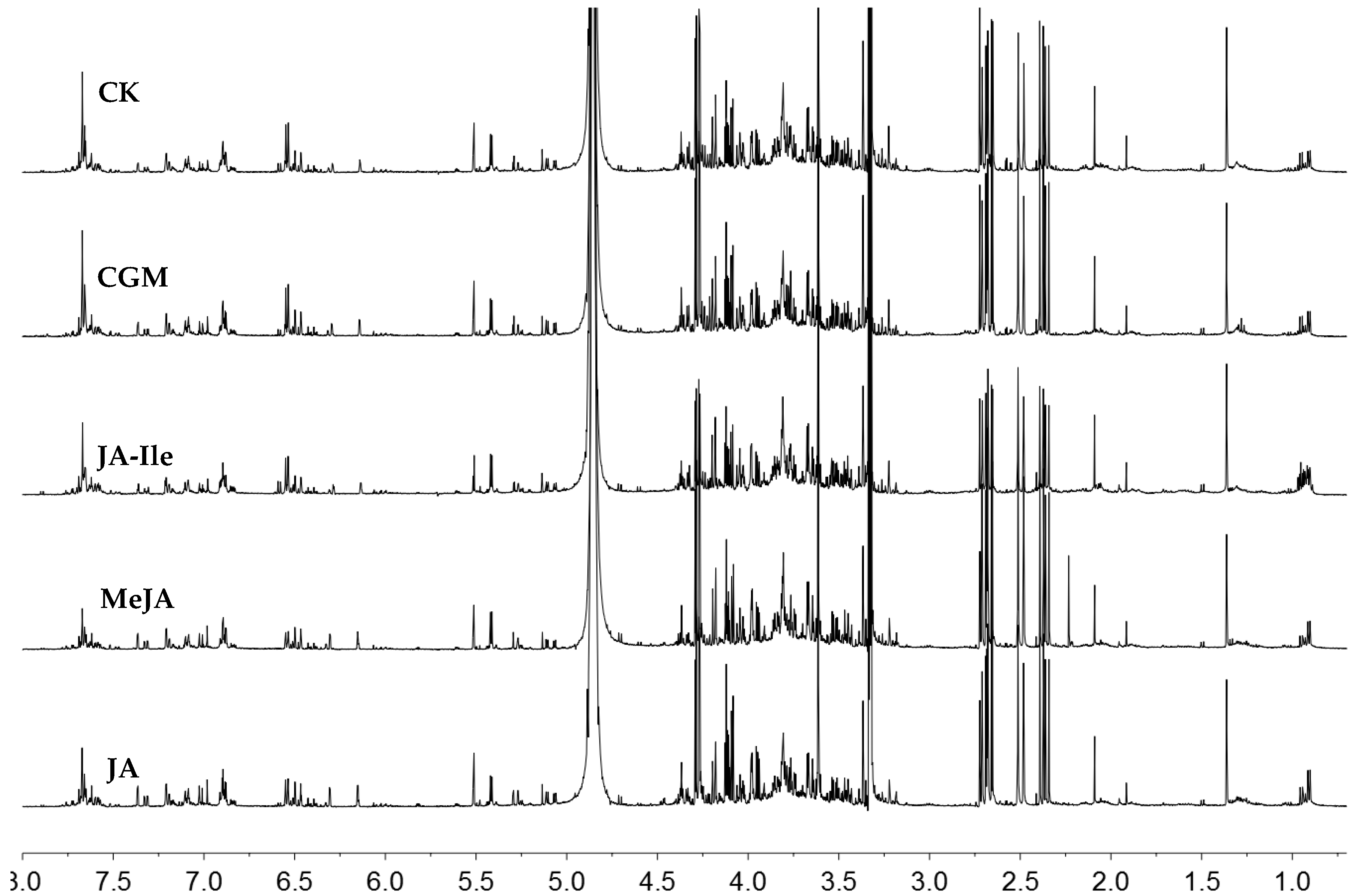
| No. | Metabolites | δH | δC |
|---|---|---|---|
| 1 | malic acid | 2.67 (dd, J = 15.3, 3.0 Hz), 2.37 (dd, J = 15.3, 10.0 Hz), 4.28 (dd, J = 10.0, 3.0 Hz) | |
| 2 | citric acid | 2.50 (d, J = 15.6 Hz), 2.69 (d, J = 15.6 Hz) | 45.7, 75.1, 179.0, 182.0 |
| 3 | formic acid | 8.46 (s) | |
| 4 | succinic acid | 2.41 (s) | |
| 5 | RC(OH)CH3-COOH | 1.36 (s) | 77.0, 180.9 |
| 6 | lactic acid | 1.34 (d, J = 6.8 Hz) | |
| 7 | fumaric acid | 6.50 (s) | |
| 8 | sucrose | 5.40 (d, J = 3.8 Hz), 4.17 (d, J = 8.7 Hz) | |
| 9 | α-glucose | 5.21 (d, J = 3.8 Hz) | 92.6 |
| 10 | β-glucose | 4.60 (d, J = 7.9 Hz) | 96.3 |
| 11 | fructose | 4.09 (1H, d, J = 3.5 Hz) | 77.6 |
| 12 | alanine | 1.49 (d, J = 7.3 Hz) | 175.5, 50.5 |
| 13 | threonine | 1.32 (d, J = 6.6 Hz) | |
| 14 | mimosine | 7.65 (overlapped, 2H), 6.54 (d, J = 6.7 Hz), 4.34 (dd, J = 14.2, 5.0 Hz), 4.23 (dd, J = 14.2, 6.7 Hz) | |
| 15 | quercetin | 7.51 (d, J = 2.1 Hz), 7.47 (dd, J = 8.6, 2.1 Hz), 6.89 (d, J = 8.5 Hz), 6.31 (d, J = 1.8 Hz), 6.13 (d, J = 1.8 Hz) | |
| 16 | quercetin-3-O-α-rhamnoside | 7.36 (d, J = 2.1 Hz), 7.32 (dd, J = 8.4, 2.1 Hz), 6.91 (1H, d, J = 8.4 Hz), 6.29 (d, J = 1.9 Hz), 6.13 (d, J = 1.9 Hz), 0.94 (d, J = 6.2) | |
| 17 | choline | 3.22 (s) | |
| 18 | steroids | 0.95 (d, J = 6.6 Hz), 0.92 (d, J = 6.7 Hz), 0.91 (d, J = 6.1 Hz), 0.95 (t, J = 7.5 Hz), 0.90 (t, J = 7.4 Hz) | 134.2 |
| 19 | 3,4-dihydroxypyridine | 7.69 (overlapped, 2H), 6.60 (d, J = 7.2 Hz) | |
| 20 | unidentified | 2.09 (s) | |
| 21 | unidentified | 2.72 (s) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Tao, Z.; Jin, Y.; Chen, S.; Zhou, Z.; Gong, A.G.W.; Yuan, Y.; Dong, T.T.X.; Tsim, K.W.K. Jasmonate-Elicited Stress Induces Metabolic Change in the Leaves of Leucaena leucocephala. Molecules 2018, 23, 188. https://doi.org/10.3390/molecules23020188
Xu Y, Tao Z, Jin Y, Chen S, Zhou Z, Gong AGW, Yuan Y, Dong TTX, Tsim KWK. Jasmonate-Elicited Stress Induces Metabolic Change in the Leaves of Leucaena leucocephala. Molecules. 2018; 23(2):188. https://doi.org/10.3390/molecules23020188
Chicago/Turabian StyleXu, Yingchao, Zhenru Tao, Yu Jin, Shuangyan Chen, Zhongyu Zhou, Amy G. W. Gong, Yunfei Yuan, Tina T. X. Dong, and Karl W. K. Tsim. 2018. "Jasmonate-Elicited Stress Induces Metabolic Change in the Leaves of Leucaena leucocephala" Molecules 23, no. 2: 188. https://doi.org/10.3390/molecules23020188




