Studies on the Chemical Diversities of Secondary Metabolites Produced by Neosartorya fischeri via the OSMAC Method
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedure
4.2. Fungus and Culture Conditions
4.3. Extraction and Isolation
4.4. Bioassay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef] [PubMed]
- Ozcengiz, G.; Demain, A.L. Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol. Adv. 2013, 31, 287–311. [Google Scholar] [CrossRef] [PubMed]
- Mulder, K.C.L.; Mulinari, F.; Franco, O.L.; Soares, M.F.; Magalhães, B.S.; Parachin, N.S. Lovastatin production: From molecular basis to industrial process optimization. Biotechnol. Adv. 2015, 33, 648–665. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem. 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Zarins-Tutt, J.S.; Barberi, T.T.; Gao, H.; Mearns-Spragg, A.; Zhang, L.X.; Newman, D.J.; Goss, R.J.M. Prospecting for new bacterial metabolites: A glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2016, 33, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Girardin, H.; Monod, M.; Latgé, J.P. Molecular characterization of the food-borne fungus Neosartorya fischeri (Malloch and Cain). Appl. Environ. Microb. 1995, 61, 1378–1383. [Google Scholar]
- Frisvad, J.C.; Larsen, T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Front. Microbiol. 2016, 6, 1485–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoler, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [PubMed]
- Zheng, Z.Z.; Shan, W.G.; Wang, S.L.; Ying, Y.M.; Ma, L.F.; Zhan, Z.J. Three new prenylated diketopiperazines from Neosartorya fischeri. Helv. Chim. Acta 2014, 97, 1020–1026. [Google Scholar] [CrossRef]
- Chen, B.Y.; Wang, Z.; Ying, Y.M.; Jiang, L.X.; Zhan, Z.J.; Wang, J.L.; Zhang, W.; Neofipiperzine, D. A new prenylated indole alkaloid metabolite of the fungus Neosartorya fischeri. J. Chem. Res. 2014, 38, 539–541. [Google Scholar]
- Shan, W.G.; Wang, S.L.; Ying, Y.M.; Ma, L.F.; Zhan, Z.J. Indole-benzodiazepine-2,5-dione derivatives from Neosartorya fischeri. J. Chem. Res. 2014, 38, 692–694. [Google Scholar] [CrossRef]
- Shan, W.G.; Wang, S.L.; Lang, H.Y.; Chen, S.M.; Ying, Y.M.; Zhan, Z.J. Cottoquinazolines E and F from Neosartorya fischeri NRRL 181. Helv. Chim. Acta 2015, 98, 552–556. [Google Scholar] [CrossRef]
- Ohshiro, T.; Rudel, L.L.; Ōmura, S.; Tomoda, H. Selectivity of microbial acyl-CoA: Cholesterol acyltransferase inhibitors toward isozymes. J. Antibiot. 2007, 60, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, R.; Goto, K.; Mitomi, M.; Oyama, K.; Sunazuka, T.; Ōmura, S. Identification of pyripyropene A as a promising insecticidal compound in a microbial metabolite screening. J. Antibiot. 2017, 70, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.J.; Fu, S.J.; Xu, M.Y.; Liang, W.L.; Lam, C.K.; Zhong, G.H.; Xu, J.; Yang, D.P.; Li, H.J. Five new cytotoxic metabolites from the marine fungus Neosartoya pseudofischri. Mar. Drugs 2016, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Ōmura, S.; Tomoda, H.; Kim, Y.K.; Nishida, H. Pyripyropenes, highly potent inhibitors of acyl-CoA:cholesterol acyltransferase produced by Aspergillus fumigatus. J. Antibiot. 1993, 46, 1168–1169. [Google Scholar] [CrossRef] [PubMed]
- Amagata, T.; Tanaka, M.; Yamada, T.; Doi, M.; Minoura, K.; Ohishi, H.; Yamori, T.; Numata, A. Variation in cytostatic constituents of a sponge-derived Gymnascella dankaliensis by manipulating the carbon source. J. Nat. Prod. 2007, 70, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wu, J.; Dai, H.F.; Xie, X.C.; Mei, W.L. Chemical constituents from Cephalotaxus endophytic fungus S26 of hainanensis. Chin. J. Med. Chem. 2008, 18, 279–283. [Google Scholar]
- Piccialli, V.; Sica, D. Four new trihydroxylated sterols from the sponge Spongionella gracillis. J. Nat. Prod. 1987, 50, 915–920. [Google Scholar] [CrossRef]
- Xiong, H.Y.; Fei, D.Q.; Zhou, J.S.; Yang, C.J.; Ma, G.L. Steroids and other constituents from the mushroom Armillaria lueovirens. Chem. Nat. Compd. 2009, 45, 759–761. [Google Scholar] [CrossRef]
- Ishizuka, T.; Yaoita, Y.; Kikuchi, M. Sterol constituents from the fruit bodies of Grifola frondosa (Fr.) S. F. Gray. Chem. Pharm. Bull. 1997, 45, 1756–1760. [Google Scholar] [CrossRef]
- Yang, S.P.; Xu, J.; Yue, J.M. Sterols from the fungus Catathelasma imperiale. Chin. J. Chem. 2003, 21, 1390–1394. [Google Scholar] [CrossRef]
- Huang, H.C.; Liaw, C.C.; Yang, H.L.; Hseu, Y.C.; Kuo, H.T.; Tsai, Y.C.; Chien, S.C.; Amagaya, S.; Chen, Y.C.; Kuo, Y.H. Lanostane triterpenoids and sterols from Antrodia camphorate. Phytochemistry 2012, 84, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Wang, H.Q.; Chen, R.Y. Studies on the constituents of the mycelia produced from fermented culture of Flammulina velutipes. Int. J. Med. Mushrooms 2003, 5, 391–396. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, Q.; Zhang, A.L.; Gao, J.M. Metabolites from Aspergillus fumigatus, an endophytic fungus associated with melia azedarach, and their antifungal, antifeedant and toxic activities. J. Agric. Food Chem. 2012, 60, 3424–3431. [Google Scholar] [CrossRef] [PubMed]
- Afiyatullov, S.S.; Kalinovskii, A.I.; Pivkin, M.V.; Dmitrenok, P.S.; Kuznetsova, T.A. Fumitremorgins from the marine isolate of the fungus Aspergillus fumigatus. Chem. Nat. Compd. 2004, 40, 615–617. [Google Scholar] [CrossRef]
- Yamazaki, M.; Fujimoto, H.; Kawasaki, T. Chemistry of tremorogenic metabolites. І. Fumitremorgin A from Aspergillus fumigatus. Chem. Pharm. Bull. 1980, 28, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Hamasaki, T.; Nakajima, H.; Isogai, A. Structure of aszonalenin, a new metabolite of Aspergillus zonatus. Tetrahedron Lett. 1982, 23, 225–228. [Google Scholar] [CrossRef]
- Tang, L.; Fu, L.L.; Lu, C.H.; Hou, X.R.; Shan, W.G.; Zhan, Z.J. New cytotoxic phloroglucinol derivatives from Agrimonia pilosa. Fitoterapia 2017, 118, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, R.; Ma, L.F.; Wo, L.K.; Hu, Y.Y.; Chen, C.; Zhan, Z.J. Aurovertin-type polyketides from Calcarrisporium arbuscular with potent cytotoxic activities against triple-negative breast cancer. Helv. Chim. Acta 2016, 99, 543–546. [Google Scholar] [CrossRef]
- Tomoda, H.; Tabata, N.; Yang, D.J.; Takayanagi, H.; Nishida, H.; Ōmura, S. Pyripyropenes, novel ACAT inhibitors produced by Aspergillus fumigatus Ⅲ. Structure elucidation of pyripyropenes E. to L. J. Antibiot. 1995, 48, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Tomoda, H.; Tabata, N.; Yang, D.J.; Namatame, I.; Tanaka, H.; Ōmura, S. Pyripyropenes, novel ACAT inhibitors produced by Aspergillus fumigatus Ⅳ. Structure elucidation of pyripyropenes M. to R. J. Antibiot. 1996, 49, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Prompanya, C.; Dethoup, T.; Bessa, L.J.; Pinto, M.M.M.; Gales, L.; Costa, P.M.; Silva, A.M.S.; Kijjoa, A. New isocoumarin derivatives and meroterpenoids from the marine sponge-associated fungus Aspergillus similanensis sp. Nov. KUFA 0013. Mar. Drugs 2014, 12, 5160–5173. [Google Scholar] [PubMed]
- Prompanya, C.; Fernandes, C.; Cravo, S.; Pinto, M.M.M.; Dethoup, T.; Silva, A.M.S.; Kijjoa, A. A new cyclic hexapeptide and a new isocoumarin derivative from the marine sponge-associated fungus Aspergillus similanensis sp. Nov. KUFA 0013. Mar. Drugs 2015, 13, 1432–1450. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.X.; Wei, J.H.; Deng, R.; Feng, G.K.; Zhu, X.F.; Lan, W.J.; Li, H.J. Two new pyripyropenes from the marine fungus Fusarium lateritium 2016F18-1. Chem. Biodivers. 2017, 14, e1600298. [Google Scholar] [CrossRef] [PubMed]
- Ohshiro, T.; Ohtawa, M.; Nagamitsu, T.; Matsuda, D.; Yagyu, H.; Davis, M.A.; Rudel, L.L.; Ishibashi, S.; Tomaoda, H. New pyripyropene A. derivatives, highly SOAT2-selective inhibitors, improve hypercholesterolemia and atherosclerosis in athrogenic mouse models. J. Pharmacol. Exp. Ther. 2015, 355, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Fuse, S.; Matsumura, K.; Johmoto, K.; Uekusa, H.; Tanaka, H.; Hirose, T.; Sunazuka, T.; Ōmura, S.; Takahashi, T. The design, synthesis, and evaluation of 1,5,7-trisubstituted-3-pyridyl-xathones for use as insecticides starting from pyripropene A. Chem. Eur. J. 2016, 22, 18450–18455. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Horikoshi, R.; Mitomi, M.; Oyama, K.; Hirose, T.; Sunazuka, T.; Ōmura, S. Synthesis and insecticidal efficacy of pyripyropene derivatives focusing on the C-1, C-7, and C-11 positions’ substituent groups. J. Antibiot. 2018, 71, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Obata, R.; Sunazuka, T.; Li, Z.R.; Tian, Z.M.; Harigaya, Y.; Tabata, N.; Tomoda, H.; Ōmura, S. Chemical modification and structure-activity relationship of pyripyropenes 1. Modification at the four hydroxyl groups. J. Antibiot. 1996, 49, 1133–1148. [Google Scholar] [CrossRef] [PubMed]
- Obata, R.; Sunazuka, T.; Harigaya, Y.; Hayashi, M.; Rho, M.C.; Tomoda, H.; Ōmura, S. Structure-activity relationships study of pyripyropenes: Reversal of cancer cell multidrug resistance. J. Antibiot. 2000, 53, 422–425. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Tokunaga, K.; Matsuda, Y.; Fujii, I.; Abe, I.; Ebizuka, Y.; Kushiro, T. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclase. Nat. Chem. 2010, 2, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Okawa, H.; Yamamoto, K.; Oyama, K.; Mitomi, M.; Anzai, H. Characterization of two cytochrome P450 monoxygenase genes of the pyripyropene biosynthetic gene cluster from Penicillium coprobium. J. Antibiot. 2011, 64, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Furutani, A.; Yamamoto, K.; Oyama, K.; Mitomi, M.; Anzai, H. Characterization of two acetyltransferase genes in the pyripyropene biosynthetic gene cluster from Penicillium coprobium. Biotechnol. Biotec. Equip. 2014, 28, 818–826. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–15 are available from the authors. |

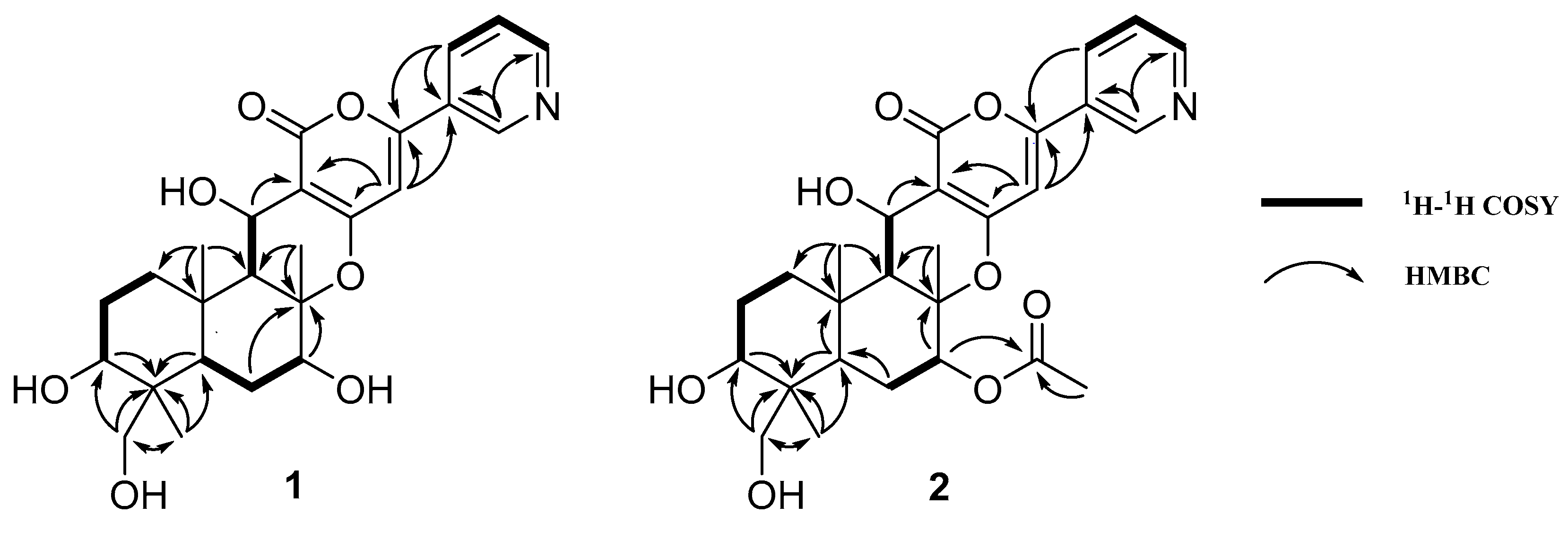
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 1 | 3.71 (dd, 12.0, 5.0) | 72.9 | 3.73 (dd, 12.0, 5.0) | 72.9 |
| 2 | 1.80 (m) | 27.3 | 1.72 (m) | 27.2 |
| 1.90 (m) | 1.93 (m) | |||
| 3 | 1.34 (m) | 37.7 | 1.37 (m) | 37.5 |
| 2.16 (m) | 2.17 (m) | |||
| 4 | - | 39.2 | - | 39.1 |
| 5 | 1.44 (d, 4.0) | 55.7 | 1.47 (d, 4.0) | 55.8 |
| 6 | - | 87.0 | - | 84.9 |
| 7 | 3.83 (dd, 11.5, 5.0) | 78.4 | 5.08 (dd, 11.5, 5.0) | 80.1 |
| 8 | 1.63 (m) | 28.9 | 1.44 (m) | 26.5 |
| 1.90 (m) | 1.84 (m) | |||
| 9 | 1.51 (dd, 12.5, 1.5) | 46.4 | 1.53 (d, 2.0) | 46.3 |
| 10 | - | 43.3 | - | 43.4 |
| 11 | 3.35 (d, 11.0) | 66.5 | 3.31 (d, 11.0) | 66.7 |
| 3.59 (d, 11.0) | 3.56 (d, 11.0) | |||
| 12 | 1.45 (s) | 18.0 | 1.45 (s) | 17.9 |
| 13 | 5.01 (d, 4.0) | 60.5 | 5.02 (d, 3.5) | 60.3 |
| 14 | 1.69 (s) | 16.0 | 1.79 (s) | 17.0 |
| 15 | 0.79 (s) | 12.7 | 0.79 (s) | 12.6 |
| 16 | - | - | - | 172.2 |
| 17 | - | - | 2.19 (s) | 21.2 |
| 18 | - | - | - | - |
| 19 | - | - | - | - |
| 2′ | - | 165.4 | - | 165.2 |
| 3′ | - | 104.4 | - | 104.5 |
| 4′ | - | 164.7 | - | 164.2 |
| 5′ | 6.86 (s) | 101.2 | 6.84 (s) | 101.1 |
| 6′ | - | 158.1 | - | 158.2 |
| 2″ | 9.07 (brs) | 147.4 | 9.08 (d, 2.0) | 147.4 |
| 3″ | - | 129.3 | - | 129.2 |
| 4″ | 8.32 (ddd, 8.0, 2.0, 1.5) | 134.9 | 8.33 (ddd, 8.0, 2.0, 1.5) | 134.9 |
| 5″ | 7.62 (dd, 8.0, 5.0) | 125.5 | 7.60 (dd, 8.0, 5.0) | 125.4 |
| 6″ | 8.69 (brd, 3.5) | 151.9 | 8.67 (dd, 5.0, 2.0) | 151.9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, Y.-M.; Huang, L.; Tian, T.; Li, C.-Y.; Wang, S.-L.; Ma, L.-F.; Shan, W.-G.; Wang, J.-W.; Zhan, Z.-J. Studies on the Chemical Diversities of Secondary Metabolites Produced by Neosartorya fischeri via the OSMAC Method. Molecules 2018, 23, 2772. https://doi.org/10.3390/molecules23112772
Ying Y-M, Huang L, Tian T, Li C-Y, Wang S-L, Ma L-F, Shan W-G, Wang J-W, Zhan Z-J. Studies on the Chemical Diversities of Secondary Metabolites Produced by Neosartorya fischeri via the OSMAC Method. Molecules. 2018; 23(11):2772. https://doi.org/10.3390/molecules23112772
Chicago/Turabian StyleYing, You-Min, Lu Huang, Ting Tian, Cui-Yu Li, Shi-Lei Wang, Lie-Feng Ma, Wei-Guang Shan, Jian-Wei Wang, and Zha-Jun Zhan. 2018. "Studies on the Chemical Diversities of Secondary Metabolites Produced by Neosartorya fischeri via the OSMAC Method" Molecules 23, no. 11: 2772. https://doi.org/10.3390/molecules23112772




