Microbial Sterolomics as a Chemical Biology Tool
Abstract
:1. Introduction
2. Phylogenic and Ecological Insights
2.1. Algal Phytosterol Biosynthesis
2.2. Trophic and Limnological Sterols
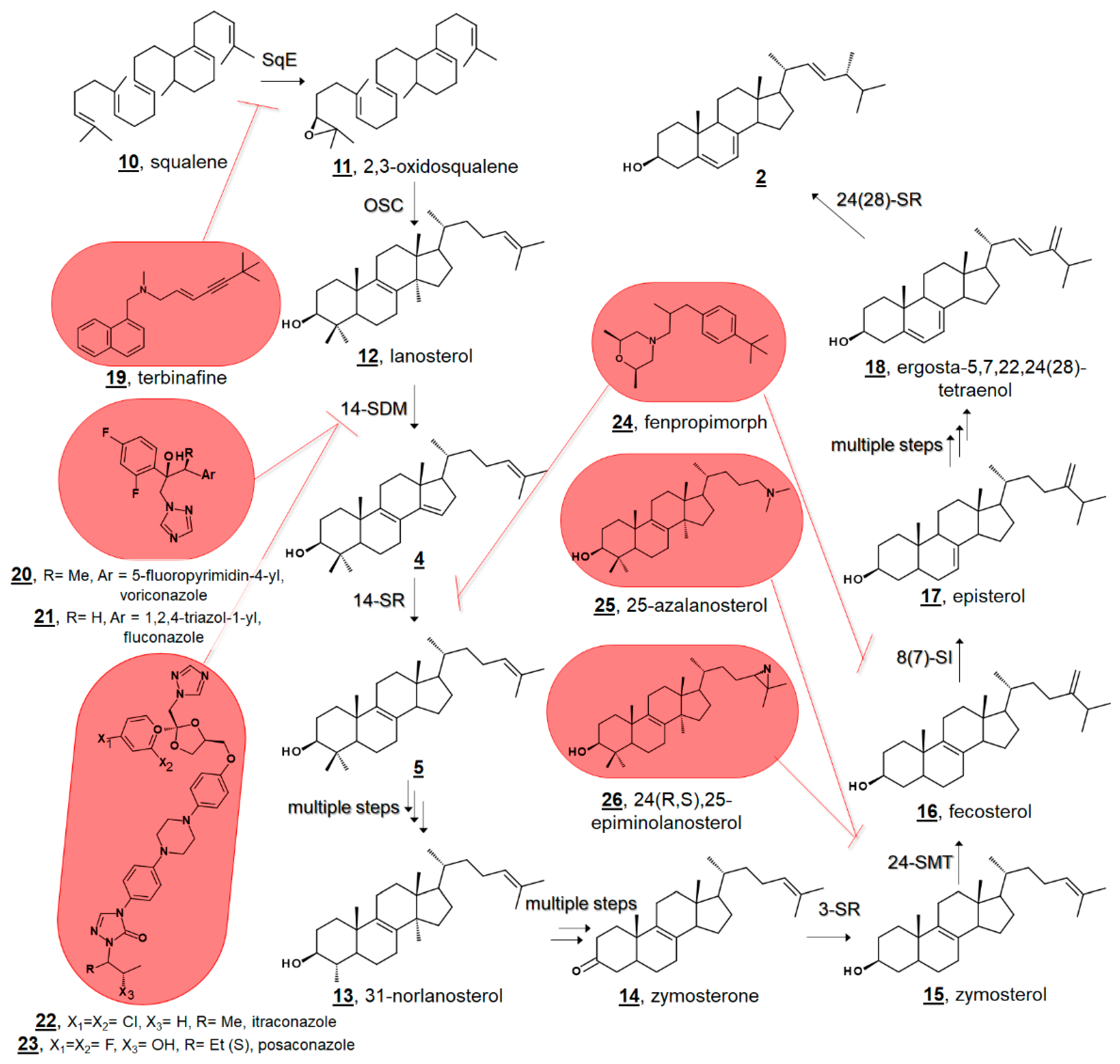
3. Sterolome-Informed Antimicrobial Targets
3.1. Trypanosoma Brucei
3.2. Acanthamoeba spp.
3.3. Fungal Sterol Profiles in Drug-Treated Cultures
4. Bioactive Steroidal Metabolites
4.1. Peroxides
4.2. Acetates
4.3. Cyclopropanes
4.4. Other Bioactive Steroids
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| [2H3]AdoMet | methyl-trideuterated S-adenosylmethionine |
| [2H3]Met | methyl-trideuterated methionine |
| 8(7)-SI | sterol C8(7)-isomerase |
| 14-SDM | sterol C14-demethylase |
| 14-SR | sterol C14-sterol reductase |
| 24-SMT | sterol C24-methyltransferase |
| Aβ | amyloid-β |
| AchE | α-acetylcholinesterase |
| BSF | bloodstream form |
| EBIs | ergosterol biosynthesis inhibitors |
| FF-MAS | follicular fluid meiosis-activating sterol |
| HEK | human epithelial kidney |
| HMGR | 3-hydroxy-3-methylglutaryl CoA reductase |
| MASs | meiosis-activating sterols |
| T-MAS | testicular meiosis-activating sterol |
| OSC | oxidosqualene cyclase |
| p38MAPK | p38 mitogen-activated protein kinase |
| PCF | procyclic form |
| PPARγ | peroxisome proliferator-activated receptor γ |
| PTP1B | protein tyrosine phosphatase 1B |
| ROCK | Rho-associated coiled-coil containing kinase |
| SqE | squalene epoxidase; TMS, trimethylsilyl(ated) |
| ZOI | zone of inhibition |
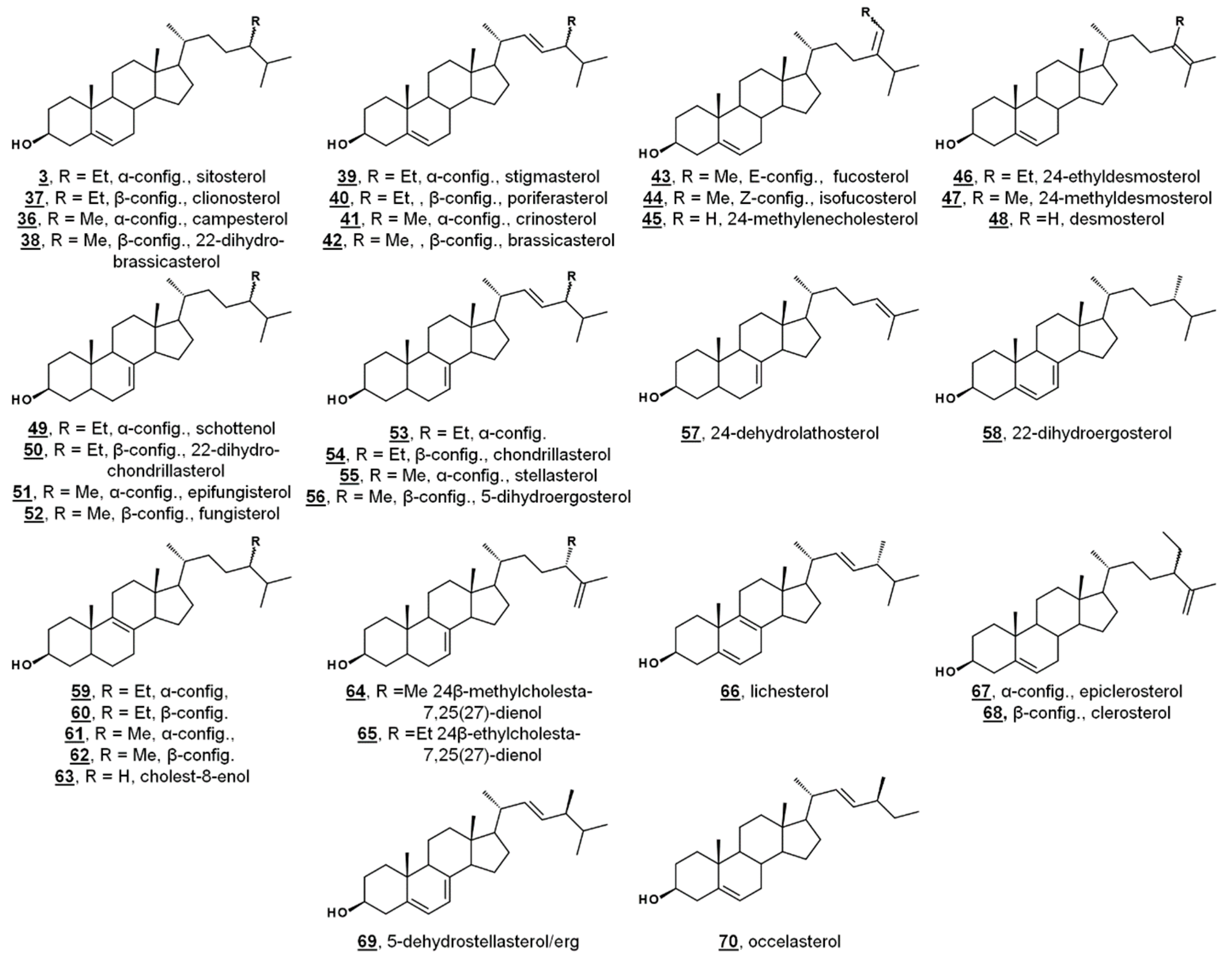
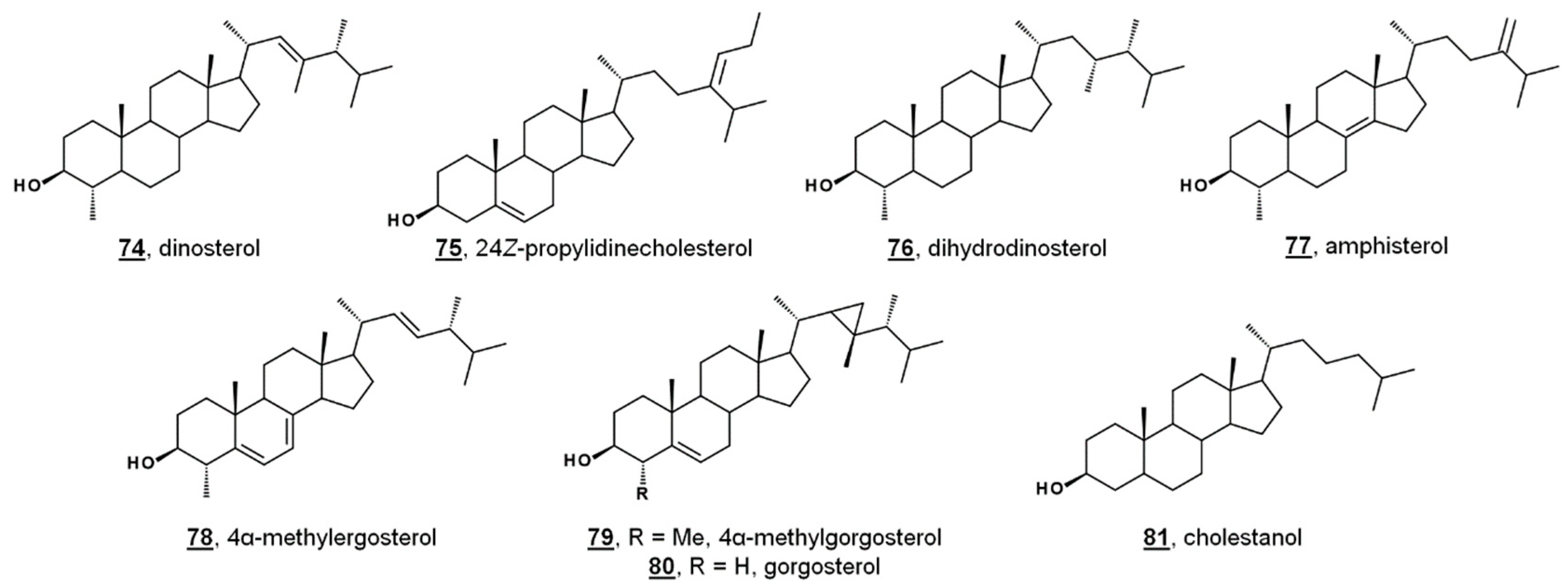
Appendix A
| No.1 | Trivial Name, If Applicable (Secondary Trivial Name, If Applicable) | Systematic Name 2 (Systematic Name Relative to 5α-Cholestane) | PubChem CID |
|---|---|---|---|
| 1 | cholesterol | cholest-5-en-3β-ol | 5997 |
| 2 | ergosterol | ergosta-5,7,22E-trien-3β-ol (24β-methylcholesta-5,7,22E-trien-3β-ol) | 444679 |
| 3 | sitosterol | stigmast-5-en-3β-ol (24α-methylcholest-5-en-3β-ol) | 222284 |
| 4 | FF-MAS | 4α,4β-dimethylcholesta-8,14,24-trien-3β-ol | 443212 |
| 5 | T-MAS | 4α,4β-dimethylcholesta-8,24-dien-3β-ol | 50990081 |
| 6 | 25-hydroxycholesterol | cholest-5-en-3β,25-diol | 65094 |
| 8 | ergosterol peroxide | 5α,8α-epidioxyergosta-6,22E-dien-3β-ol (24β-methyl-5α,8α-epidioxycholesta-6,22E-dien-3β-ol) | 5351516 |
| 9 | squalamine | 3β-[3-(4-aminobutyl)amino]propyl-7α-hydroxycholestan-24β-hydrosulfate | 72495 |
| 12 | lanosterol | lanosta-8,24-dien-3β-ol (4α,4β,14α-trimethylcholesta-8,24-dien-3β-ol) | 246983 |
| 13 | 31-norlanosterol | 4α,14α-dimethylcholesta-8,24-dien-3β-ol | 15101557 |
| 14 | zymosterone | cholesta-8,24-dien-3-one | 22298942 |
| 15 | zymosterol | cholesta-8,24-dien-3β-ol | 92746 |
| 16 | fecosterol | ergosta-8,24(28)-dien-3β-ol (24-methylideneholest-8-en-3β-ol) | 440371 |
| 17 | episterol | ergosta-7,24(28)-dien-3β-ol (24-methylideneholest-7-en-3β-ol) | 5283662 |
| 18 | ergosta-5,7,22E,24(28)-tetraen-3β-ol (24-methylideneholesta-5,7,22E-trien-3β-ol) | 11090531 | |
| 25 | 25-azalanosterol | 25-azalanost-8(9)-en-3β-ol 4α,4β,14α-trimethyl-25-azacholest-8-en-3β-ol | 66746490 |
| 26 | 24(R,S),25-epiminolanosterol | 24(R,S),25-epiminolanost8(9)-en-3β-ol (4α,4β,14α-trimethyl-24,25-azanetriylcholest-8-en-3β-ol) | 163740 |
| 27 | cycloartenol | cycloart-24(25)-en-3β-ol (4α,4β,14α-trimethyl-9β,19-cyclocholest-24-en-3β-ol) | 92110 |
| 28 | cyclolaudenol | 24β-methylcycloart-25(27)-en-3β-ol (4α,4β,14α,24β-tetramethyl-9β,19-cyclocholest-25(27)-en-3β-ol) | 101729 |
| 29 | 24-methylenecycloartanol | 24-methylidenecycloartan-3β-ol (4α,4β,14α-trimethyl-24-methylidene-9β,19-cyclocholestan-3β-ol) | 94204 |
| 30 | ergosta-8,25(27)-dien-3β-ol (24β-methylcholest-8,25(27)-dien-3β-ol) | 102515129 | |
| 31 | obtusifoliol | 4α,14α-dimethylergosta-8,24(28)-dien-3β-ol (4α,14α-dimethyl-24-methylideneholest-8-en-3β-ol) | 65252 |
| 32 | 24(28)-methylenelophenol | 4α-methylergosta-7,24(28)-dien-3β-ol (4α-methyl-24-methylideneholest-7-en-3β-ol) | 5283640 |
| 33 | chlamysterol | 4α,14α-dimethylporiferasta-8,25(27)-dien-3β-ol (24β-ethyl-4α,14α-dimethyl-cholesta-8,25(27)-dien-3β-ol) | 90657605 |
| 34 | 24(28)Z-ethylidene lophenol (citrostadienol) | 4α-methylstigmasta-7,24(28)Z-dien-3β-ol (4α-methyl-24Z-ethylideneholest-7-en-3β-ol) | 9548595 |
| 35 | 7-dehydroporiferasterol | poriferasta-5,7,22E-trien-3β-ol (24β-ethylcholesta-5,7,22E-trien-3β-ol) | 20843308 |
| 36 | campesterol | campest-5-en-3β-ol (24α-methylcholest-5-en-3β-ol) | 173183 |
| 37 | clionosterol (22-dihydroporiferasterol) | poriferast-5-en-3β-ol (24β-ethylcholest-5-en-3β-ol) | 457801 |
| 38 | 22-dihydrobrassicasterol | ergost-5-en-3β-ol (24β-methylcholesta-5-en-3β-ol) | 312822 |
| 39 | stigmasterol | stigmasta-5,22E-dien-3β-ol (24α-ethylcholesta-5,22E-dien-3β-ol) | 5280794 |
| 40 | poriferasterol | poriferasta-5,22E-dien-3β-ol (24β-ethylcholesta-5,22E-dien-3β-ol) | 5281330 |
| 41 | crinosterol (epibrassiasterol) | campesta-5,22E-dien-3β-ol (24α-methylcholesta-5,22E-dien-3β-ol) | 5283660 |
| 42 | brassicasterol | ergosta-5,22E-3β-ol (24β-methylcholesta-5,22E-dien-3β-ol) | 5281327 |
| 43 | fucosterol | stigmasta-5,24(28)E-dien-3β-ol (24E-ethylideneholest-7-en-3β-ol) | 5281328 |
| 44 | isofucosterol | stigmasta-5,24(28)Z-dien-3β-ol (24Z-ethylideneholest-5-en-3β-ol) | 5281326 |
| 45 | 24(28)-methylenecholesterol (chalinasterol) | ergosta-5,24(28)-dien-3β-ol (24-methylideneholest-5-en-3β-ol) | 92113 |
| 46 | 24-ethyldesmosterol | stigmasta-5,24(25)-dien-3β-ol (24-ethylcholesta-5,24(25)-dien-3β-ol) | 22848721 |
| 47 | 24-methyldesmosterol | ergosta-5,24(25)-dien-3β-ol (24-methylcholesta-5,24(25)-dien-3β-ol) | 193567 |
| 48 | desmosterol | cholesta-5,24-dien-3β-ol | 439577 |
| 49 | schottenol | stigmast-7-en-3β-ol (24α-ethylcholest-7-en-3β-ol) | 441837 |
| 50 | 22-dihydrochondrillasterol | poriferast-7-en-3β-ol (24β-ethylcholest-7-en-3β-ol) | 5283639 |
| 51 | epifungisterol (22-dihydrostellasterol) | campest-7-en-3β-ol (24α-methylcholest-7-en-3β-ol) | 90889779 |
| 52 | fungisterol | ergost-7-en-3β-ol (24β-methylcholest-7-en-3β-ol) | 5283646 |
| 53 | stigmasta-7,22E-dien-3β-ol (24α-ethylcholesta-7,22E-dien-3β-ol) | 125122456 | |
| 54 | chondrillasterol | poriferasta-7,22E-dien-3β-ol (24β-ethylcholesta-7,22E-dien-3β-ol) | 5283663 |
| 55 | stellasterol | campesta-7,22E-dien-3β-ol (24α-methylcholest-7,22E-dien-3β-ol) | 5283669 |
| 56 | 5-dihydroergosterol | ergosta-7,22E-dien-3β-ol (24β-methylcholest-7,22E-dien-3β-ol) | 13889661 |
| 57 | 24-dehydrolathosterol | cholesta-7,24-dien-3β-ol | 5459827 |
| 58 | 22-dihydroergosterol | ergosta-5,7-dien-3β-ol (24β-methylcholest-5,7-dien-3β-ol) | 5326970 |
| 59 | stigmast-8-en-3β-ol (24α-ethylcholest-8-en-3β-ol) | 23424905 | |
| 60 | poriferast-8-en-3β-ol (24β-ethylcholest-8-en-3β-ol) | 101826503 | |
| 61 | campest-8-en-3β-ol (24α-methylcholest-8-en-3β-ol) | - | |
| 62 | ergost-8-en-3β-ol (24β-methylcholest-8-en-3β-ol) | 60077053 | |
| 63 | cholest-8-enol (24-dihydrozymosterol) | cholest-8-en-3β-ol | 101770 |
| 64 | ergosta-7,25(27)-dien-3β-ol (24β-methylcholesta-7,25(27)-dien-3β-ol) | 60077052 | |
| 65 | poriferasta-7,25(27)-dien-3β-ol (24β-ethylcholesta-7,25(27)-dien-3β-ol) | 5283655 | |
| 66 | lichesterol | ergosta-5,8,22E-trien-3β-ol (24β-methylcholesta-5,8,22E-trien-3β-ol) | 5281329 |
| 67 | clerosterol | poriferasta-5,25(27)-dien-3β-ol (24β-ethylcholesta-5,25(27)-dien-3β-ol) | 5283638 |
| 68 | epiclerosterol | stigmasta-5,25(27)-dien-3β-ol (24α-ethylcholesta-5,25(27)-dien-3β-ol) | 185472 |
| 69 | 5-dehydrostellasterol (epiergosterol) | campesta-5,7,22E-trien-3β-ol (24α-methylcholesta-5,7,22E-trien-3β-ol) | 124427258 |
| 70 | occelasterol | 27-norcholesta-5,22E-dien-3β-ol | 15481847 |
| 71 | lathosterol | cholest-7-en-3β-ol | 65728 |
| 72 | 7-dehydrocholesterol (provitamin D3) | cholesta-5,7-dien-3β-ol | 439423 |
| 73 | fucosteryl epoxide | 24,28-epoxyergost-5-en-3β-ol 24-(3-methyloxiran-2-yl)-cholest-5-en-3β-ol | 3082427 |
| 74 | dinosterol | 4α,23-dimethylergost-22E-en-3β-ol (4α,23,24β-trimethylcholest-22E-en-3β-ol) | 44263330 |
| 75 | 24Z-propylidenecholesterol | 6443745 | |
| 76 | dihydrodinosterol | (23R)-4α,23-dimethylergostan-3β-ol ((23R)-4α,23,24β-trimethylcholestan-3β-ol) | 133309 |
| 77 | amphisterol | 4α-methylergosta-8(14),24(28)-dien-3β-ol (4α-methyl-24-methylidenechoest-8(14)-en-3β-ol) | 60077061 |
| 78 | 4α-methylergosterol | 4α-methylergosta-5,7,22E-trien-3β-ol (4α,24β-dimethylcholesta-5,7,22E-trien-3β-ol) | - |
| 79 | 4α-methylgorgosterol | 4α-methylgorgost-5-en-3β-ol ((22R,23R)-4α,23,24β-trimethyl-22,23-methanocholest-5-en-3β-ol) | - |
| 80 | gorgosterol | gorgost-5-en-3β-ol ((22R,23R)-23,24β-dimethyl-22,23-methanocholest-5-en-3β-ol) | 52931413 |
| 81 | cholestanol | (5α) cholestan-3β-ol | 6710664 |
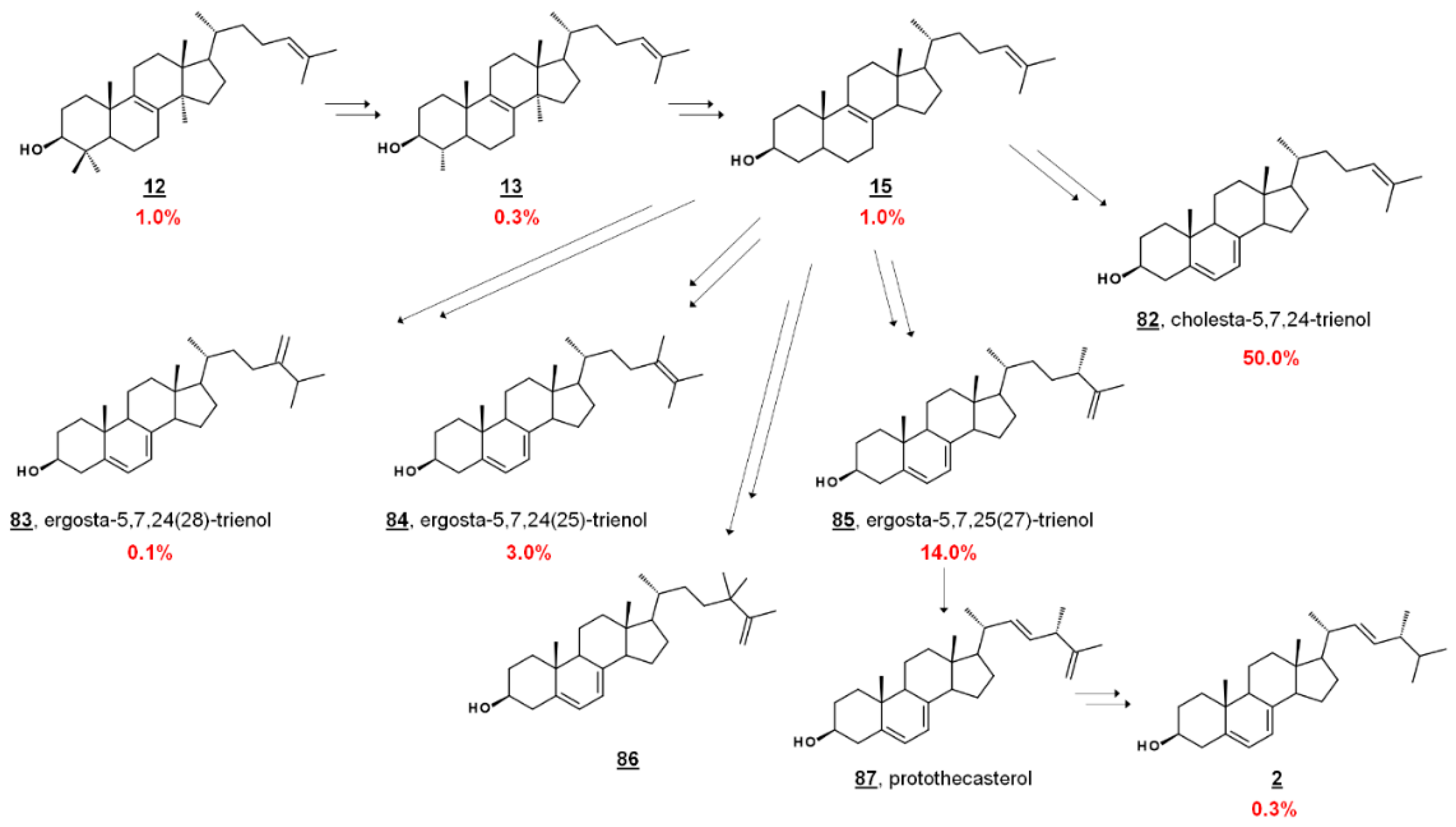
| 82 | cholesta-5,7,24-trienol 7-dehydrodesmosterol | cholesta-5,7,24-trien-3β-ol | 440558 |
| 83 | ergosta-5,7,24(28)-trien-3β-ol (24β-methylcholesta-5,7,24(28)-trien-3β-ol) | 10894570 | |
| 84 | ergosta-5,7,24(25)-trien-3β-ol (24β-methylcholesta-5,7,24(25)-trien-3β-ol) | 58104987 | |
| 85 | ergosta-5,7,25(27)-trien-3β-ol (24β-methylcholesta-5,7,25(27)-trien-3β-ol) | 101600336 | |
| 86 | 24,24-dimethylcholesta-5,7,25(27)-trien-3β-ol | - | |
| 87 | protothecasterol | ergosta-5,7,22E,25(27)-tetraen-3β-ol (24β-methylcholesta-5,7,22E,25(27)-tetraen-3β-ol) | 101600338 |
| 88 | 26-fluorolanosterol | 26-fluorolanosta-8,24-dien-3β-ol (26-fluoro-4α,4β,14α-trimethylcholesta-8,24-dien-3β-ol) | - |
| 89 | 26-fluorocholesta-5,7,24-trien-3β-ol | - | |
| 90 | 26-fluoro-4α,4β-dimethylcholesta-8,24-dien-3β-ol | - | |
| 91 | 26-fluoro-4α-methylcholesta-8,24-dien-3β-ol | - | |
| 92 | 26-fluorocholesta-8,24-dien-3β-ol | - | |
| 93 | 26-fluoroergosta-8,25(27)-dien-3β-ol 26-fluoro-24β-methylcholesta-8,25(27)-dien-3β-ol | - | |
| 94 | amebasterol-1 | 19(10→6)-abeo-ergosta-5,7,9,22E-tetraen-3β-ol (24β-methyl-19(10→6)-abeo-cholesta-5,7,9,22E-tetraen-3β-ol) | 11596359 |
| 95 | amebasterol-2 | 19(10→6)-abeo-poriferasta-5,7,9,22E-tetraen-3β-ol (24β-ethyl-19(10→6)-abeo-cholesta-5,7,9,22E-tetraen-3β-ol) | - |
| 96 | amebasterol-3 | 19(10→6)-abeo-ergosta-5,7,9-trien-3β-ol (24β-methyl-19(10→6)-abeo-ergosta-5,7,9-trien-3β-ol) | - |
| 97 | amebasterol-4 | 19(10→6)-abeo-poriferasta-5,7,9,22E,25-pentaen-3β-ol (24β-ethyl-19(10→6)-abeo-cholesta-5,7,9,22E,25-pentaen-3β-ol) | - |
| 98 | amebasterol-5 | 19(10→6)-abeo-poriferasta-5,7,9,25(27)-tetraen-3β-ol (24β-ethyl-19(10→6)-abeo-cholesta-5,7,9,25(27)-tetraen-3β-ol) | - |
| 99 | amebasterol-6 | 19(10→6)-abeo-poriferasta-5,7,9-trien-3β-ol (24β-ethyl-19(10→6)-abeo-poriferasta-5,7,9-trien-3β-ol) | - |
| 102 | eburicol | 24-methylidenelanost-8-en-3β-ol 4α,4β,14α-trimethyl-24-methylidenecholest-8-en-3β-ol | 9803310 |
| 103 | 14-methylergosta-8,24(28)-dien-3β,6α-diol 14,24β-dimethyl-24-methylidenecholest-8-en--3β,6α-diol | 148910 | |
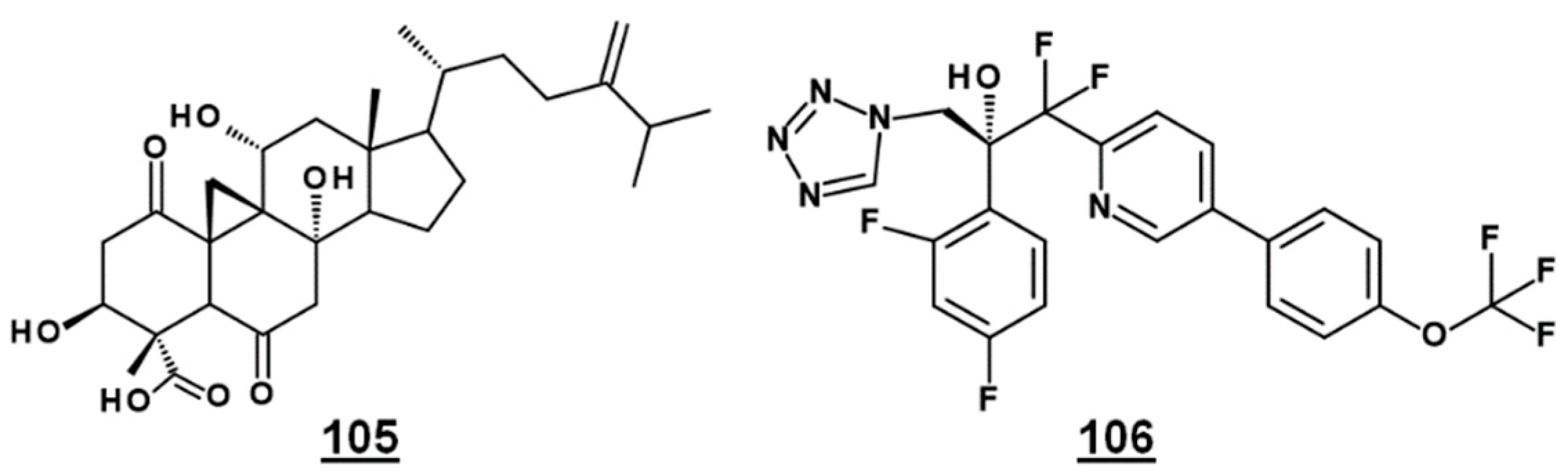
| 105 | FR171456 | 24-methylidene-3β,8α,11α-trihydroxy-1,6-dioxocycloartan-30-oic acid (4α,14α-dimethyl-24-methylidene-9β,19-cyclo-3β,8α,11α-trihydroxy-1,6-dioxocholestan-4β-carboxylic acid) | - |
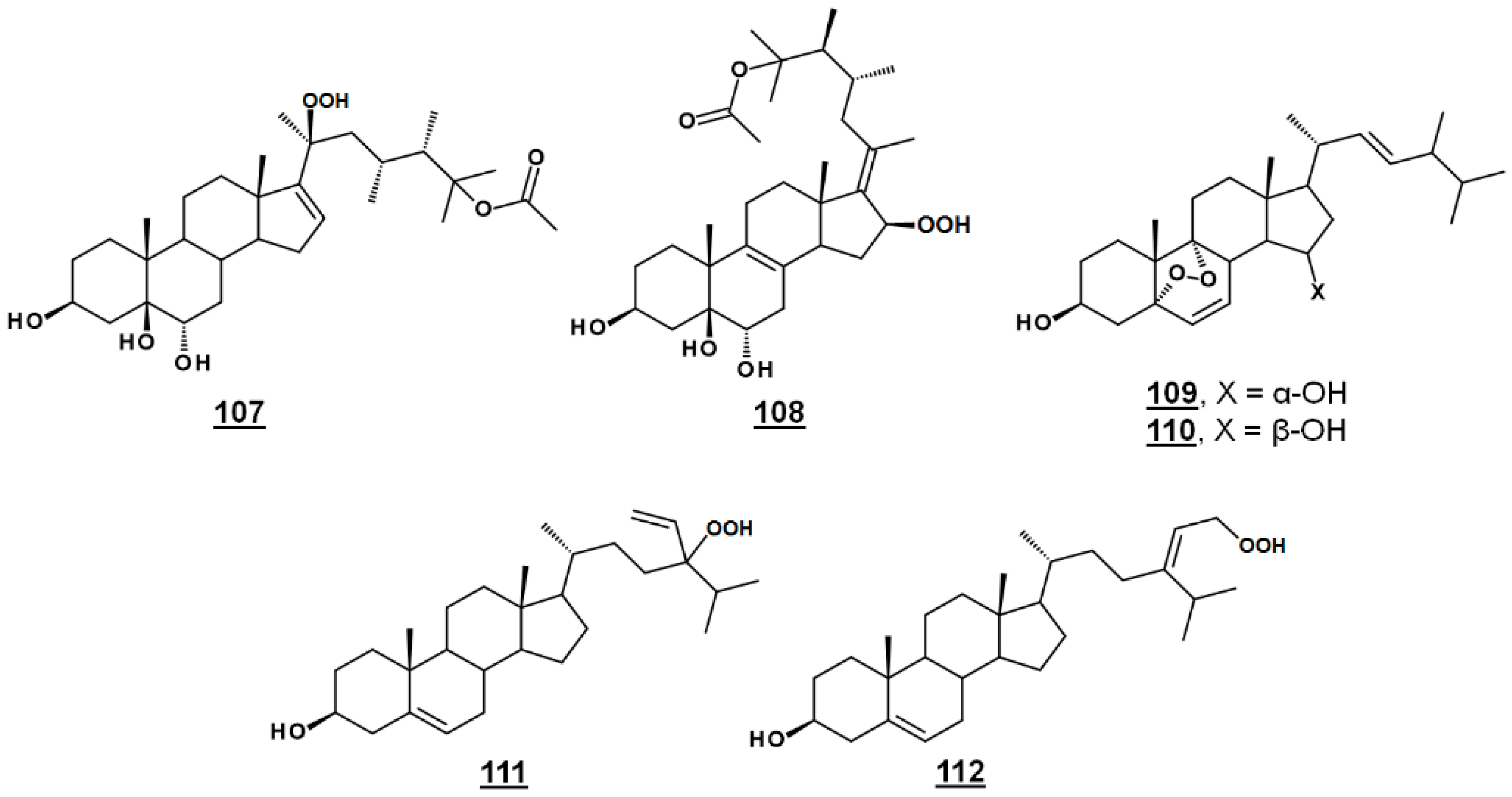
| 107 | michosterol A | (20S,23R)-23-methyl-20-hydroperoxy-25-acetoxyergost-16-en-3β,5β,6α-triol ((20S,23R)-23,24β-dimethyl-20-hydroperoxy-25-acetoxycholest-16-en-3β,5β,6α-triol) | - |
| 108 | michosterol B | (17E,23R)-23-methyl-16-hydroperoxy-25-acetoxyergost-17-en-3β,5β,6α-triol ((17E,23R)-23,24β-di-methyl-16-hydroperoxy-25-acetoxycholest-17-en-3β,5β,6α-triol) | - |
| 109 | nigerasterol A | 5α,9α-epidioxyergosta-6,8(14),22E-trien-3β,15α-diol (24β-methyl-5α,9α-epidioxycholesta-6,8(14),22E-trien-3β,15α-diol) | - |
| 110 | nigerasterol B | 5α,9α-epidioxyergosta-6,8(14),22E-trien-3β,15β-diol (24β-methyl-5α,9α-epidioxycholesta-6,8(14),22E-trien-3β,15β-diol) | - |
| 111 | 24-ethenyl-24-hydroperoxycholest-5-en-3β-ol | 10411225 | |
| 112 | 29-hydroperoxyisofucosterol | (24Z)-29-hydroperoxystigmasta-5,24(28)-dien-3β-ol 24Z-(2-hydroperoxyethylidnene)cholest-5-en-3β-ol | 46224335 |
| 113 | michosterol C | 6α-acetoxyergostan-3β,5β,25-triol (24β-methyl-6α-acetoxycholestan-3β,5β,25-triol) | - |
| 114 | anicequol (NGA0187) | 16β-acetoxy-3β,7β,11β-trihydroxyergost-22E-en-6-one (24β-methyl-16β-acetoxy-3β,7β,11β-trihydroxycholest-22E-en-6-one) | 10413810 |
| 115 | penicisteroid A | 24-methyl-16β-acetoxycholest-22E-en-3β,6β,7β,11β-tetrol | - |
| 116 | penicisteroid C | 24-methyl-16β-acetoxycholesta-5,22E-dien-3β,6β-diol | - |
| 117 | 11α-acetoxycholest-24-en-3β,5α,6β-triol | - | |
| 118 | 11α-acetoxyergosta-22E,25-dien-3β,5α,6β-triol 24β-methyl-11α-acetoxycholesta-22E,25-dien-3β,5α,6β-triol | - | |
| 119 | 11α-acetoxygorgostan-3β,5α,6β-triol ((22R,23R)-23,24β-dimethyl-22,23-methano-11α-acetoxycholestan-3β,5α,6β-triol) | 54769262 | |
| 120 | halicrasterol D | 11α-acetoxyergost-22E-en-3β,5α,6β-triol 24β-methyl-11α-acetoxycholest-22E-en-3β,5α,6β-triol | - |
| 121 | 11α,19-diacetoxycholest-7-en-2α,3β,5α,6β,9α-pentol | - | |
| 122 | 6β-acetoxyergost-24(28)-en-3β,5α-diol 24-methylidene-6β-acetoxycholestan -3β,5α-diol | 101687891 | |
| 123 | methyl 25-acetoxy-3β-hydroxycholest-5-en-19-carboxylate | - | |
| 124 | 11α-acetoxygorgostan-3β,5α,6β,12α-tetrol ((22R,23R)-23,24β-dimethyl-22,23-methano-11α-acetoxycholest-5-en-3β,5α,6β,12α-tetrol) | 56962930 | |
| 125 | 12α-acetoxygorgostan-3β,5α,6β,11α-tetrol ((22R,23R)-23,24β-dimethyl-22,23-methano-12α-acetoxycholestan-3β,5α,6β,11α-tetrol) | - | |
| 126 | 11α-acetoxygorgostan-3β,5α,6β,15α-tetrol ((22R,23R)-23,24β-dimethyl-22,23-methano-12α-acetoxycholestan-3β,5α,6β,15α-tetrol) | - | |
| 127 | 7β-acetoxyergosta-5,24(28)-dien-3β,19-diol 24-methylidene-7β-acetoxycholest-5-en-3β,19-diol | 477494 | |
| 128 | halymeniaol | 3β,15α,16β-triacetoxy-12β-hydroxycholest-5-en-7-one | - |
| 129 | 21-O-octadecanoyl-xestokerol A | (20S,21R)-21-octadecanoyl-11β,20,22-trihydroxypetrostan-3-one ((20S,21R,25R,26R)-24α,26-dimethyl-26,27-cyclo-21-octadecanoyl-11β,20,22-trihydroxycholestan-3-one) | 71747680 |
| 130 | xestokerol A | (20S,21R)-11β,20,21,22-tetrahydroxypetrostan-3-one ((20S,21R,25R,26R)-24α,26-dimethyl-26,27-cyclo-11β,20,21,22-tetrahydroxycholestan-3-one) | 44584465 |
| 131 | xestokerol A dimethyl ketal | (20S,21R)-3,3-dimethoxypetrostan-11β,20,21,22-tetrol ((20S,21R,25R,26R)-24α,26-dimethyl-26,27-cyclo-3,3-dimethoxycholestan--11β,20,21,22-tetrol) | - |
| 132 | 7α-hydroxypetrosterol | petrost-5-en-3β,7α-diol ((25R,26R)-24α,26-dimethyl-26,27-cyclocholest-5-en-3β,7α-diol) | 101209535 |
| 133 | 7β-hydroxypetrosterol | petrost-5-en-3β,7β-diol ((25R,26R)-24α,26-dimethyl-26,27-cyclocholest-5-en-3β,7β-diol) | 71747681 |
| 134 | 7-ketopetrosterol | 3β-hydroxypetrost-5-en-7-one ((25R,26R)-24α,26-dimethyl-26,27-cyclo-3β-hydroxycholest-5-en-7-one) | 101209534 |
| 135 | petrosterol | petrost-5-en-3β-ol ((25R,26R)-24α,26-dimethyl-26,27-cyclocholest-5-en-3β-ol) | 194249 |
| 136 | 11β-hydroxypetrosterol | petrost-5-en-3β,11βα-diol ((25R,26R)-24α,26-dimethyl-26,27-cyclocholest-5-en-3β,11β-diol) | - |
| 137 | (20S)-petrostan-3α,7α,12β,20-tetrol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclocholestan-3α,7α,12β,20-tetrol) | - | |
| 138 | (20S)-petrostan-3α,12β,14α,20-tetrol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclocholestan-3α,12β,14α,20-tetrol) | - | |
| 139 | (20S)-3,3-dimethoxypetrostan-7α,12β,20-triol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclo-3,3-dimethoxycholestan-7α,12β,20-triol) | - | |
| 140 | (20S)-3,3-dimethoxypetrostan-7α,12β,19,20-tetrol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclo-3,3-dimethoxycholestan-7α,12β,19,20-tetrol) | - | |
| 141 | (20S)-petrostan-3α,12β,20-triol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclocholestan-3α,12β,20-triol) | - | |
| 142 | (20S)-petrostan-3β,12β,20-triol ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclocholestan-3β,12β,20-triol) | - | |
| 143 | aragusterol B | (20S)-12β,20-dihydroxypetrostan-3-one ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclo-12β,20-dihydroxycholestan-3-one) | 44566420 |
| 144 | (20S)-7α,12β,20-trihydroxypetrostan-3-one ((20S,25R,26R)-24α,26-dimethyl-26,27-cyclo-7α,12β,20-trihydroxycholestan-3-one) | - | |
| 145 | 3,3-dimethoxypetrostan-12β,16α-diol ((25R,26R)-24α,26-dimethyl-26,27-cyclo-3,3-dimethoxycholestan-12β,16α-diol) | - | |
| 146 | aragusterol A | (20R,22S)-20,21-epoxy-12β,22-dihydroxypetrostan-3-one ((20R,22S,25R,26R)-24α,26-dimethyl-20,21-epoxy-26,27-cyclo-12β,22-dihydroxycholestan-3-one) | 9933873 |
| 147 | (20R,22S)-20,21-epoxy-3,3-dimethoxypetrostan-12β,22-diol ((20R,22S,25R,26R)-24α,26-dimethyl-20,21-epoxy-26,27-cyclo-3,3-dimethoxycholestan-12β,22-diol) | 10696885 | |
| 148 | (22R)-12β,22-dihydroxypetrost-20(21)-en-3-one ((22R,25R,26R)-24α,26-dimethyl-26,27-cyclo-12β,22-dihydroxycholest-20(21)-en--3-one) | - | |
| 149 | aragusterol J | (22R)-7β,12β,22-trihydroxypetrost-20(21)-en-3-one ((22R,25R,26R)-24α,26-dimethyl-26,27-cyclo-7β,12β,22-trihydroxycholest-20(21)-en--3-one) | - |
| 150 | klyflaccisteroid C | 3β,7α-dihydroxygorgost-5-en-11-one ((22R,23R)-23,24β-dimethyl-22,23-methano-3β,7α-dihyroxycholest-5-en-11-one) | - |
| 151 | klyflaccisteroid D | 3β-hydroxygorgost-5-en-7,11-dione ((22R,23R)-23,24β-dimethyl-22,23-methano-3β,7α-dihyroxycholest-5-en-7,11-dione) | - |
| 152 | klyflaccisteroid E | gorgosta-5,9(11)-dien-3β,7β,12α-triol ((22R,23R)-23,24β-dimethyl-22,23-methanocholesta-5,9(11)-dien-3β,7β,12α-triol) | - |
| 153 | gorgost-5-en-3β,9α,11α-triol ((22R,23R)-23,24β-dimethyl-22,23-methanocholest-5,-dien-3β,9α,11α-triol) | 10742556 | |
| 154 | klyfaccisteroid H | gorgost-5-en-3β,11α,12α-triol ((22R,23R)-23,24β-dimethyl-22,23-methanocholesta-5,9(11)-dien-3β,11α,12α-triol) | - |
| 155 | halistanol sulfate | 24,25-dimethylcholestane-2β,3α,6α-trisulfate | 73361 |
| 156 | halistanol sulfate I | 24-methyl-24,25-methanocholestane-2β,3α,6α-trisulfate | - |
| 157 | halistanol sulfate J | 24,24-(methylethano)cholestane-2β,3α,6α-trisulfate | - |
| 158 | solomonsterol A | trisodium cholane-2β,3α,24-trisulfate (trisodium 25,26,27-trinorcholestane-2β,3α,24-trisulfate) | 50925451 |
| 159 | solomonsterol B | trisodium 24-norcholane-2β,3α,23-trisulfate (trisodium 24,25,26,27-tetranorcholestane-2β,3α,24-trisulfate) | 53318073 |
| 160 | theonellasterol | 4-methylidineporiferast-8(14)-en-3β-ol (24β-ethyl-4-methylidinecholest-8(14)-en-3β-ol) | 52931395 |
| 161 | conicasterol | 4-methylidinecampest-8(14)-en-3β-ol (24α-methyl-4-methylidinecholest-8(14)-en-3β-ol) | 21670674 |
| 162 | ganoderic acid A | 7β,15α-dihydroxy-3,11,23-trioxolanost-8-en-26-oic acid 4α,4β,14α-trimethyl-7β,15α-dihydroxy-3,11,23-trioxocholest-8-en-26-oic acid) | 471002 |
| 163 | ergosta-7,9(11),22E-trien-3β-ol 24β-methylcholesta-7,9(11),22E-trien-3β-ol | 12308954 | |
| 164 | ergosta-4,7,22E-trien-3-one 24β-methylcholesta-4,7,22E-trien-3-one | 11003773 | |
| 165 | ergosta-4,6,8(14),22E-tetraen-3-one 24β-methylcholesta-4,6,8(14),22E-tetraen-3-one | 6441416 | |
| 166 | 14α-hydroxyergosta-4,7,9(11),22E-tetraen-3,6-dione 24β-methyl-14α-hydroxycholesta-4,7,9(11),22E-tetraen-3,6-dione | 10251684 | |
| 167 | 9α,14α-dihydroxyergosta-4,7,22E-trien-3,6-dione 24β-methyl-9α,14α-dihydroxycholesta-4,7,22E-trien-3,6-dione | - | |
| 168 | ergosta-4,6,8(14),22E,24(28)-pentaen-3-one 24-methylidenecholesta-4,6,8(14),22E-tetraen-3-one | - | |
| 169 | nodulisporiviridin E | 18-nor-1α,3β-dihydroxy-4,5,6-[2,3,4]furanoandrosta-5,8,11,13(14)-tetraen-7,17-dione 18,20,21,22,23,24,25,26,27-nonanor-1α,3β-dihydroxy-4,5,6-[2,3,4]furanoandrosta-5,8,11,13(14)-tetraen-7,17-dione | 122179368 |
| 170 | nodulisporiviridin F | 3β,11β-dihydroxy-4,5,6-[2,3,4]furanoandrosta-5,8-dien-7,17-dione 20,21,22,23,24,25,26,27-octanor-3β,11β-dihydroxy-4,5,6-[2,3,4]furanocholesta-5,8-dien-7,17-dione | 122179369 |
| 171 | nodulisporiviridin G | 11β-hydroxy-4,5,6-[2,3,4]furanoandrosta-5,8-dien-3,7,17-trione 20,21,22,23,24,25,26,27-octanor-11β-hydroxy-4,5,6-[2,3,4]furanocholesta-5,8-dien-3,7,17-trione | 122179370 |
| 172 | nodulisporiviridin H | 3β,12β-dihydroxy-4,5,6-[2,3,4]furanoandrosta-5,8-dien-7,17-dione 20,21,22,23,24,25,26,27-octanor-3β,12β-dihydroxy-4,5,6-[2,3,4]furanocholesta-5,8-dien-7,17-dione | 122179371 |
| 173 | 16-O-desmethylasporyergosterol-β-d-mannoside | β-d-mannosyloxyergosta-6,8(14),17(20)E, 22E-tetraen-3β-ol 24β-methyl-β-d-mannosyloxycholesta-6,8(14),17(20)E,22E-tetraen-3β-ol | - |
| 174 | (24S)-24,28-epoxyergost-5-en-3β,4α-diol (24S)-24-oxyranylcholest-5-en-3β,4α-diol | 44575614 | |
| 175 | ergosta-5,24(28)-dien-3β,7α-diol (24-methylidenecholest-5-en-3β,7α-diol) | 10949727 | |
| 176 | ergosta-5,24(28)-dien-3β,7β-diol (24-methylidenecholest-5-en-3β,7β-diol) | 11373355 | |
| 177 | ergost-5-en-3β,7β-diol (24β-methylcholest-5-en-3β,7β-diol) | 11475561 | |
| 178 | ergost-24(28)-en-3β,5α,6β-triol (24-methylidenecholestan-3β,5α,6β-triol) | 21775108 | |
| 179 | ergostan-3β,5α,6β-triol (24β-methylcholestan-3β,5α,6β-triol) | 44558918 | |
| 180 | 3β,5α,6β,11α-tetrahydroxyergostan-1-one (24β-methyl-3β,5α,6β,11α-tetrahydroxycholestan-1-one) | - | |
| 181 | ergostan-1α,3β,5α,6β,11α-pentol (24β-methylcholestan-1α,3β,5α,6β,11α-pentol) | - | |
| 182 | sarcoaldesterol B | ergostan-3β,5α,6β,11α-tetrol (24β-methylcholestan-3β,5α,6β,11α-tetrol) | 10718409 |
| 183 | ergostan-1β,3β,5α,6β-tetrol (24β-methylcholestan-1β,3β,5α,6β-tetrol) | - | |
| 184 | pregnedioside A | 4α-O-β-d-arabinopyranosyloxypregn-20-en-3β-ol22,23,24,25,26,27-hexanor-4α-O-β-d-arabinopyranosyloxycholest-20-en-3β-ol | 21673267 |
| 185 | gorgostan-1α,3β,5α,6β,11α-pentol ((22R,23R)-23,24β-dimethyl-22,23-methanocholestan-1α,3β,5α,6β,11α-pentol) | 23426029 | |
| 186 | sarcoaldesterol A | gorgostan-3β,5α,6β,11α-tetrol ((22R,23R)-23,24β-dimethyl-22,23-methanocholestan-3β,5α,6β,11α-tetrol) | 10790775 |
| 187 | (20R,23R)-23-methylergost-16-en-3β,20-diol (20R,23R)-23,24β-methylcholest-16-en-3β,20-diol | - | |
| 188 | ximaosteroid E | (16S)-16,22-epoxycholesta-1,22E-dien-3-one | - |
| 189 | ximaosteroid F | (20R,22R)-20,22-dihydroxycholesta-1,4-dien-3-one | - |
| 190 | (20S)-20-hydroxycholest-1-en-3,16-dione | 53997071 | |
| 191 | sinubrasone A | methyl (22R)-22-O-β-d-xylopyranosyloxy-3-oxoergosta-1,4-diene-26-carboxylate methyl (22R)-24β-methyl-22-O-β-d-xylopyranosyloxy-3-oxocholesta-1,4-diene-26-carboxylate | - |
| 192 | sinubrasone B | methyl (16S,22R)-16-methoxy-16,22-epoxy-3-oxoergosta-1,4-diene-26-carboxylate methyl (16S,22R)-24β-methyl-16-methoxy-16,22-epoxy-3-oxocholesta-1,4-diene-26-carboxylate | - |
| 193 | sinubrasone C | methyl (22R,23R)-22,23-epoxy-3-oxoergosta-1,4-diene-26-carboxylate methyl (22R,23R)-24β-methyl-22,23-epoxy-3-oxocholesta-1,4-diene-26-carboxylate | - |
| 194 | sinubrasone D | methyl (20S)-20-methyl-3-oxopregna-1,4-diene-21-carboxylate methyl 23,24,25,26,27-pentanor-3-oxocholesta-1,4-diene-21-carboxylate | 15929041 |
| 195 | ergostan-1α,3β,5α,6β,11α,15α-hexol (24β-methylcholestan-1α,3β,5α,6β,11α,15α-hexol) | - | |
| 196 | ergostan-3β,5α,6β,15α-tetrol (24β-methylcholestan-3β,5α,6β,15α-tetrol) | - | |
| 197 | ergostan-3β,5α,6β,11α,15α-pentol (24β-methylcholestan-3β,5α,6β,11α,15α-pentol) | - | |
| 198 | ergost-7-en-3β,5α,6β,15α-tetrol (24β-methylcholest-7-en-3β,5α,6β,15α-tetrol) | - | |
| 199 | 23-methylergost-22E-en-3β,5α,6β,11α-tetrol (23,24β-dimethylcholest-22E-en-3β,5α,6β,11α-tetrol) | - | |
| 200 | klyflaccisteroid A | (17S,23R)-23-methylergosta-5,20(21)-dien-3β,17α-diol (17S,23R)-23,24β-dimethylcholesta-5,20(21)-dien-3β,17α-diol | - |
| 201 | klyfaccisteroid J | (20R,23R)-23-methylergosta-5,16-dien-3β,11α,20-triol (20R,23R)-23,24β-dimethylcholesta-5,16-dien-3β,11α,20-triol | - |
| 202 | klyflaccisteroid M | (22S)-ergost-5-en-3β,7β,22-triol ((22S)-24β-methylcholest-5-en-3β,7β,22-triol) | - |
| 203 | subergorgol U | 19(10→4)-abeo-2-hydroxypregna-2,4,1(10)-trien-20-one (22,23,24,25,26,27-hexnor-19(10→4)-abeo-2-hydroxycholesta-2,4,1(10)-trien-20-one) | 132918691 |
| 204 | 19(10→4)-abeo-1-hydroxypregna-2,4,1(10)-trien-20-one (22,23,24,25,26,27-hexnor-19(10→4)-abeo-2-hydroxycholesta-2,4,1(10)-trien-20-one) | 54484024 | |
| 205 | (20S)-7α,12β,20-trihydroxycholest-22E-en-3-one | - | |
| 206 | langcosterol A | 26,27-dimethylergosta-5,24(28)-dien-3β,7α-diol (26,27-dimethyl-24-methylidenecholest-5-en-3β,7α-diol) | 23426186 |
| 207 | ergosta-4,7,22E,25-tetraen-3-one (24β-methylcholesta-4,7,22E,25-tetraen-3-one) | 132280531 | |
| 208 | 7α,12β,18-trihydoxystigmast-22E-en-3-one (24α-methyl-7α,12β,18-trihydoxycholest-22E-en-3-one) | - | |
| 209 | (20S)-24-ethyl-7α,12β,20-trihydroxycholestan-3-one | - | |
| 210 | (20S)-24-methyl-7α,12β,20-trihydroxycholest-22E-en-3-one | - | |
| 211 | 7,15-dioxoconicasterol | 4-methylidene-3β-hydroxycampest-8(14)-en-7,15-dione (24α-methyl-4-methylidene-3β-hydroxycholest-8(14)-en-7,15-dione) | - |
| 212 | 15-oxoconicasterol | 4-methylidene-3β-hydroxycampest-8(14)-en-15-one (24α-methyl-4-methylidene-3β-hydroxycholest-8(14)-en-15-one) | - |
| 213 | 4-methylidene-3β,9α-dihydroxycampest-8(14)-en-15-one (24α-methyl-4-methylidene-3β,9α-dihydroxycholest-8(14)-en-15-one) | - | |
| 214 | gelliusterol E | 24-methylchola-5,16-dien-23-yn-3β,7α-diol (26,27-dinorcholesta-5,16-dien-23-yn-3β,7α-diol) | - |
| 215 | saringosterol | 24-ethenylcholest-5-en-3β,24-diol | 14161394 |
| 216 | dictyosterol A | 3β,6β-dihydroxycholesta-4,22E-dien-24-one | - |
| 217 | dictyosterol B | 6β-hydroxycholesta-4,22E-dien-3,24-dione | - |
| 218 | dictyosterol C | 3β,7α-dihydroxycholesta-5,22E-dien-24-one | - |
| 219 | 3β-hydroxycholesta-5,22E-dien-7,24-dione | - | |
| 220 | 3β-hydroxycholesta-5,22E-dien-24-one | - | |
| 221 | dictyopterisin C | (24R)-stigmasta-4,28(29)-dien-3β,7β,24-triol (24R)-24-ethenylcholest-4-en-3β,7β,24-triol | - |
| 222 | (24R)-7-methoxystigmasta-4,28(29)-dien-3β,24-diol (24R)-7-methoxy-24-ethenylcholest-4-en-3β,24-diol | - | |
| 223 | dictyopterisin F | (24R)-3β,24-dihydroxystigmasta-4,28(29)-dien-7-one (24R)-24-ethenyl-3β,24-dihydroxycholest-4-en-7-one | - |
| 224 | dictyopterisin G | (24S)-3β,24-dihydroxyporiferasta-4,28(29)-dien-7-one (24S)-24-ethenyl-3β,24-dihydroxycholest-4-en-7-one | - |
| 225 | dictyopterisin H | (24R)-stigmasta-4,28(29)-dien-3β,6β,24-triol (24R)-24-ethenylcholest-4-en-3β,6β,24-triol | - |
| 226 | dictyopterisin I | (24R)-6β,24-dihydroxystigmasta-4,28(29)-dien-3-one (24R)-24-ethenyl-6β,24-dihydroxycholest-4-en-3-one | - |
| 227 | dictyopterisin J | (24S)-6β,24-dihydroxyporiferasta-4,28(29)-dien-3-one (24S)-24-ethenyl-6β,24-dihydroxycholest-4-en-3-one | - |
References
- Nes, W.D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451. [Google Scholar] [CrossRef] [PubMed]
- Volkman, J.K. Sterols and other triterpenoids: Source specificity and evolution of biosynthetic pathways. Org. Geochem. 2005, 36, 139–159. [Google Scholar] [CrossRef]
- Cheng, L.L.; Wang, G.Z.; Zhang, H.Y. Sterol biosynthesis and prokaryotes-to-eukaryotes evolution. Biochem. Biophys. Res. Commun. 2007, 363, 885–888. [Google Scholar] [CrossRef] [PubMed]
- Desmond, E.; Gribaldo, S. Phylogenomics of sterol synthesis: Insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 2009, 1, 364–381. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.H.; Yin, X.; Welander, P.V. Sterol synthesis in diverse bacteria. Front. Microbiol. 2016, 7, 990. [Google Scholar] [CrossRef] [PubMed]
- Blackstone, N.W. An evolutionary framework for understanding the origin of eukaryotes. Biology 2016, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Tabas, I. Cholesterol in health and disease. J. Clin. Investig. 2002, 110, 583–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santori, F.R.; Huang, P.; van de Pavert, S.A.; Douglass, E.F., Jr.; Leaver, D.J.; Haubrich, B.A.; Keber, R.; Lorbek, G.; Konijn, T.; Rosales, B.N.; et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell. Metab. 2015, 21, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Poirot, M.; Silvente-Poirot, S. The tumor-suppressor cholesterol metabolite, dendrogenin A, is a new class of LXR modulator activating lethal autophagy in cancers. Biochem. Pharmacol. 2018, 153, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Zerbinati, C.; Caponecchia, L.; Puca, R.; Ciacciarelli, M.; Salacone, P.; Sebastianelli, A.; Pastore, A.; Palleschi, G.; Petrozza, V.; Porta, N.; et al. Mass spectrometry profiling of oxysterols in human sperm identifies 25-hydroxycholesterol as a marker of sperm function. Redox Biol. 2017, 11, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Abdel-Khalik, J.; Hearn, T.; Yutuc, E.; Morgan, A.H.; Wang, Y. Current trends in oxysterol research. Biochem. Soc. Trans. 2016, 44, 652–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Testa, G.; Rossin, D.; Poli, G.; Biasi, F.; Leonarduzzi, G. Implication of oxysterols in chronic inflammatory human diseases. Biochimie 2018. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Wang, Y. Sterolomics: State of the art, developments, limitations and challenges. Biochim. Biophys. Acta. 2017, 1862, 771–773. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Wang, Y. An update on oxysterol biochemistry: New discoveries in lipidomics. Biochem. Biophys. Res. Commun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Moreau, R.A.; Nystrom, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid. Res. 2018, 70, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Nes, W.R.; Krevitz, K.; Joseph, J.; Nes, W.D. The phylogenetic distribution of sterols in tracheophytes. Lipids 1977, 12, 511–527. [Google Scholar] [CrossRef]
- Weete, J.D. Structure and function of sterols in fungi. In Advances in Lipid Research; Paoletti, R., Kritchevsky, D., Eds.; Elsevier: New York, NY, USA, 1989; Volume 23, pp. 115–167. [Google Scholar]
- Weete, J.D.; Gandhi, S.R. Sterols of the phylum zygomycota: Phylogenetic implications. Lipids 1997, 32, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Furlong, S.T.; Samia, J.A.; Rose, R.M.; Fishman, J.A. Phytosterols are present in Pneumocystis carinii. Antimicrob. Agents Chemother. 1994, 38, 2534–2540. [Google Scholar] [CrossRef] [PubMed]
- Nes, W.D.; Norton, R.A.; Crumley, F.G.; Madigan, S.J.; Katz, E.R. Sterol phylogenesis and algal evolution. Proc. Natl. Acad. Sci. USA 1990, 87, 7565–7569. [Google Scholar] [CrossRef] [PubMed]
- Halevy, S.; Avivi, L.; Katan, H. Sterols of soil amoebas and Ochromonas danica: Phylogenetic approach. J. Protozool. 1966, 13, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Raederstorff, D.; Rohmer, M. Sterol biosynthesis de nova via cycloartenol by the soil amoeba Acanthamoeba polyphaga. Biochem. J. 1985, 231, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Burden, R.S.; Cooke, D.T.; Carter, G.A. Inhibitors of sterol biosynthesis and growth in plants and fungi. Phytochemistry 1989, 28, 1791–1804. [Google Scholar] [CrossRef]
- Odds, F.C.; Brown, A.J.; Gow, N.A. Antifungal agents: Mechanisms of action. Trends Microbiol. 2003, 11, 272–279. [Google Scholar] [CrossRef]
- Leaver, D.J. Synthesis and Biological Activity of Sterol 14alpha-Demethylase and Sterol C24-Methyltransferase Inhibitors. Molecules 2018, 23, 1753. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Nes, W.D. Steroidal triterpenes: Design of substrate-based inhibitors of ergosterol and sitosterol synthesis. Molecules 2009, 14, 4690–4706. [Google Scholar] [CrossRef] [PubMed]
- Godtfredsen, W.O.; Jahnsen, S.; Lorck, H.; Roholt, K.; Tybring, L. Fusidic acid: A new antibiotic. Nature 1962, 193, 987. [Google Scholar] [CrossRef] [PubMed]
- Verbist, L. The antimicrobial activity of fusidic acid. J. Antimicrob. Chemother. 1990, 25 (Suppl. B), 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Godecke, T.; Gunn, J.; Duan, J.A.; Che, C.T. Protostane and fusidane triterpenes: A mini-review. Molecules 2013, 18, 4054–4080. [Google Scholar] [CrossRef] [PubMed]
- Bu, M.; Yang, B.B.; Hu, L. Natural bioactive sterol 5α, 8α-endoperoxides as drug lead compounds. Med. Chem. 2014, 4, 709–716. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Bioactive fungal endoperoxides. Med. Mycol. 2015, 1, 5. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.M.; Li, C.S.; Wang, B.G. Nigerasterols A and B, antiproliferative sterols from the mangrove-derived endophytic fungus Aspergillus niger MA-132. Helvetica. Chimica. Acta 2013, 96, 1055–1061. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, J.; Ma, W.; Fang, W.; Chen, Z.; Yang, B.; Liu, Y. Bioactivities of six sterols isolated from marine invertebrates. Pharm. Biol. 2014, 52, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Kitchawalit, S.; Kanokmedhakul, K.; Kanokmedhakul, S.; Soytong, K. A new benzyl ester and ergosterol derivatives from the fungus Gymnoascus reessii. Nat. Prod. Res. 2014, 28, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Brunel, J.M.; Salmi, C.; Loncle, C.; Vidal, N.; Letourneux, Y. Squalamine: A polyvalent drug of the future? Curr. Cancer Drug Targets 2005, 5, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Warrilow, A.G.S.; Thomas, C.D.; Ramos, E.; Parker, J.E.; Price, C.L.; Vanderloop, B.H.; Fisher, P.M.; Loftis, M.D.; Kelly, D.E.; et al. Functional importance for developmental regulation of sterol biosynthesis in Acanthamoeba castellanii. Biochim. Biophys. Acta 2018, 1863, 1164–1178. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.; Rice, C.A.; Zhang, T.; Edrada-Ebel, R.; Henriquez, F.L.; Roberts, C.W. Characterisation of sterol biosynthesis and validation of 14alpha-demethylase as a drug target in Acanthamoeba. Sci. Rep. 2017, 7, 8247. [Google Scholar] [CrossRef] [PubMed]
- Korn, E.D.; Von Brand, T.; Tobie, E.J. The sterols of Trypanosoma cruzi and Crithidia fasciculata. Comp. Biochem. Physiol. 1969, 30, 601–610. [Google Scholar] [CrossRef]
- Goad, L.J.; Holz, G.G., Jr.; Beach, D.H. Sterols of Leishmania species. Implications for biosynthesis. Mol. Biochem. Parasitol. 1984, 10, 161–170. [Google Scholar] [CrossRef]
- Nakamura, C.V.; Waldow, L.; Pelegrinello, S.R.; Ueda-Nakamura, T.; Filho, B.A.; Filho, B.P. Fatty acid and sterol composition of three phytomonas species. Mem. Inst. Oswaldo Cruz. 1999, 94, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Haubrich, B.A.; Singha, U.K.; Miller, M.B.; Nes, C.R.; Anyatonwu, H.; Lecordier, L.; Patkar, P.; Leaver, D.J.; Villalta, F.; Vanhollebeke, B.; et al. Discovery of an ergosterol-signaling factor that regulates Trypanosoma brucei growth. J. Lipid Res. 2015, 56, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Nes, C.R.; Singha, U.K.; Liu, J.; Ganapathy, K.; Villalta, F.; Waterman, M.R.; Lepesheva, G.I.; Chaudhuri, M.; Nes, W.D. Novel sterol metabolic network of Trypanosoma brucei procyclic and bloodstream forms. Biochem. J. 2012, 443, 267–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Cross, G.A.; Nes, W.D. Cholesterol import fails to prevent catalyst-based inhibition of ergosterol synthesis and cell proliferation of Trypanosoma brucei. J. Lipid. Res. 2007, 48, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Urbina, J.A. Ergosterol biosynthesis and drug development for Chagas disease. Mem. Inst. Oswaldo Cruz 2009, 104 (Suppl. 1), 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.B.; Haubrich, B.A.; Wang, Q.; Snell, W.J.; Nes, W.D. Evolutionarily conserved Delta(25(27))-olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii. J. Lipid. Res. 2012, 53, 1636–1645. [Google Scholar] [CrossRef] [PubMed]
- Gealt, M.A.; Adler, J.H.; Nes, W.R. The sterols and fatty acids from purified flagella of Chlamydomonas reinhardtii. Lipids 1981, 16, 133–136. [Google Scholar] [CrossRef]
- Patterson, G.W. The distribution of sterols in algae. Lipids 1971, 6, 120–127. [Google Scholar] [CrossRef]
- Volkman, J.K. Sterols in microalgae. In The Physiology of Microalgae; Borowitzka, M., Beardall, J., Raven, J., Eds.; Springer: Cham, Switzerland, 2016; Volume 6, pp. 485–505. [Google Scholar]
- Patkar, P.; Haubrich, B.A.; Qi, M.; Nguyen, T.T.; Thomas, C.D.; Nes, W.D. C-24-methylation of 26-fluorocycloartenols by recombinant sterol C-24-methyltransferase from soybean: Evidence for channel switching and its phylogenetic implications. Biochem. J. 2013, 456, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Haubrich, B.A.; Collins, E.K.; Howard, A.L.; Wang, Q.; Snell, W.J.; Miller, M.B.; Thomas, C.D.; Pleasant, S.K.; Nes, W.D. Characterization, mutagenesis and mechanistic analysis of an ancient algal sterol C24-methyltransferase: Implications for understanding sterol evolution in the green lineage. Phytochemistry 2015, 113, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nes, W.D.; Marshall, J.A.; Jia, Z.; Jaradat, T.T.; Song, Z.; Jayasimha, P. Active site mapping and substrate channeling in the sterol methyltransferase pathway. J. Biol. Chem. 2002, 277, 42549–42556. [Google Scholar] [CrossRef] [PubMed]
- Dahmen, A.S.; Houle, H.M.; Leblond, J.D. Free sterols of the historically important green algal genera, Acetabularia and Acicularia (Polyphysaceae): A modern interpretation. Algol. Stud. 2017, 2017, 59–70. [Google Scholar] [CrossRef]
- Martin-Creuzburg, D.; Merkel, P. Sterols of freshwater microalgae: Potential implications for zooplankton nutrition. J. Plankton Res. 2016, 38, 865–877. [Google Scholar] [CrossRef]
- Giner, J.L.; Zhao, H.; Tomas, C. Sterols and fatty acids of three harmful algae previously assigned as Chattonella. Phytochemistry 2008, 69, 2167–2171. [Google Scholar] [CrossRef] [PubMed]
- Giner, J.L.; Zhao, H.; Dixon, M.S.; Wikfors, G.H. Bioconversion of (13)C-labeled microalgal phytosterols to cholesterol by the Northern Bay scallop, Argopecten irradians irradians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 192, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gergs, R.; Steinberger, N.; Beck, B.; Basen, T.; Yohannes, E.; Schulz, R.; Martin-Creuzburg, D. Compound-specific delta(13)C analyses reveal sterol metabolic constraints in an aquatic invertebrate. Rapid. Commun. Mass Spectrom. 2015, 29, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Martin-Creuzburg, D.; von Elert, E. Ecological significance of sterols in aquatic food webs. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M., Arts, M., Eds.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Behmer, S.T.; Nes, W.D. Insect sterol nutrition and physiology: A global overview. Adv. Insect. Physiol. 2003, 31, 72. [Google Scholar] [CrossRef]
- Taipale, S.J.; Hiltunen, M.; Vuorio, K.; Peltomaa, E. Suitability of Phytosterols Alongside Fatty Acids as Chemotaxonomic Biomarkers for Phytoplankton. Front. Plant. Sci. 2016, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.X.; Yu, R.C.; Chen, Z.F.; Peng, Q.C.; Yan, T.; Zhou, M.J. Analysis of sterols in selected bloom-forming algae in China. Harmful. Algae. 2017, 66, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Giner, J.L.; Ceballos, H.; Tang, Y.Z.; Gobler, C.J. Sterols and Fatty Acids of the Harmful Dinoflagellate Cochlodinium polykrikoides. Chem. Biodivers. 2016, 13, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Houle, H.M.; Lopez, C.B.; Leblond, J.D. Sterols of the Toxic Marine Dinoflagellate, Pyrodinium bahamense. J. Eukaryot. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Tolosa, I.; Orejas, C.; Rueda, L.; Viladrich, N.; Grinyo, J.; Flogel, S.; Grover, R.; Ferrier-Pages, C. Biochemical composition of the cold-water coral Dendrophyllia cornigera under contrasting productivity regimes: Insights from lipid biomarkers and compound-specific isotopes. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 2018. [Google Scholar] [CrossRef]
- Crandell, J.B.; Teece, M.A.; Estes, B.A.; Manfrino, C.; Ciesla, J.H. Nutrient acquisition strategies in mesophotic hard corals using compound specific stable isotope analysis of sterols. J. Exp. Mar. Bio. Ecol. 2016, 474, 133–141. [Google Scholar] [CrossRef]
- Zhou, W.; Lepesheva, G.I.; Waterman, M.R.; Nes, W.D. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei. J. Biol. Chem. 2006, 281, 6290–6296. [Google Scholar] [CrossRef] [PubMed]
- Millerioux, Y.; Mazet, M.; Bouyssou, G.; Allmann, S.; Kiema, T.R.; Bertiaux, E.; Fouillen, L.; Thapa, C.; Biran, M.; Plazolles, N.; et al. De novo biosynthesis of sterols and fatty acids in the Trypanosoma brucei procyclic form: Carbon source preferences and metabolic flux redistributions. PLoS Pathog. 2018, 14, e1007116. [Google Scholar] [CrossRef] [PubMed]
- Leaver, D.J.; Patkar, P.; Singha, U.K.; Miller, M.B.; Haubrich, B.A.; Chaudhuri, M.; Nes, W.D. Fluorinated Sterols Are Suicide Inhibitors of Ergosterol Biosynthesis and Growth in Trypanosoma brucei. Chem. Biol. 2015, 22, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Dauchy, F.A.; Bonhivers, M.; Landrein, N.; Dacheux, D.; Courtois, P.; Lauruol, F.; Daulouede, S.; Vincendeau, P.; Robinson, D.R. Trypanosoma brucei CYP51: Essentiality and Targeting Therapy in an Experimental Model. PLoS Negl. Trop Dis. 2016, 10, e0005125. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Ott, R.D.; Hargrove, T.Y.; Kleshchenko, Y.Y.; Schuster, I.; Nes, W.D.; Hill, G.C.; Villalta, F.; Waterman, M.R. Sterol 14alpha-demethylase as a potential target for antitrypanosomal therapy: Enzyme inhibition and parasite cell growth. Chem. Biol. 2007, 14, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Lorente, S.O.; Rodrigues, J.C.; Jimenez Jimenez, C.; Joyce-Menekse, M.; Rodrigues, C.; Croft, S.L.; Yardley, V.; de Luca-Fradley, K.; Ruiz-Perez, L.M.; Urbina, J.; et al. Novel azasterols as potential agents for treatment of leishmaniasis and trypanosomiasis. Antimicrob. Agents Chemother. 2004, 48, 2937–2950. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Patkar, P.; Singha, U.K.; Chaudhuri, M.; David Nes, W. 24-Methylenecyclopropane steroidal inhibitors: A Trojan horse in ergosterol biosynthesis that prevents growth of Trypanosoma brucei. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Kidane, M.E.; Vanderloop, B.H.; Zhou, W.; Thomas, C.D.; Ramos, E.; Singha, U.; Chaudhuri, M.; Nes, W.D. Sterol methyltransferase a target for anti-amoeba therapy: Towards transition state analog and suicide substrate drug design. J. Lipid. Res. 2017, 58, 2310–2323. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Warrilow, A.G.; Rolley, N.J.; Parker, J.E.; Nes, W.D.; Smith, S.N.; Kelly, D.E.; Kelly, S.L. Azole Antifungal Agents to Treat the Human Pathogens Acanthamoeba castellanii and Acanthamoeba polyphaga through Inhibition of Sterol 14alpha-Demethylase (CYP51). Antimicrob. Agents Chemother. 2015, 59, 4707–4713. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, H.; Tanuma, K.; Enomoto, K.; Yoshimura, Y.; Onuki, T.; Nihonyanagi, S.; Hamada, Y.; Noguchi, N. Evaluation of In Vitro Antiamoebic Activity of Antimicrobial Agents Against Clinical Acanthamoeba Isolates. J. Ocul. Pharmacol. Ther. 2017, 33, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Gueudry, J.; Le Goff, L.; Compagnon, P.; Lefevre, S.; Colasse, E.; Aknine, C.; Duval, F.; Francois, A.; Razakandrainibe, R.; Ballet, J.J.; et al. Evaluation of voriconazole anti-Acanthamoeba polyphaga in vitro activity, rat cornea penetration and efficacy against experimental rat Acanthamoeba keratitis. J. Antimicrob. Chemother. 2018. [Google Scholar] [CrossRef] [PubMed]
- Taravaud, A.; Loiseau, P.M.; Pomel, S. In vitro evaluation of antimicrobial agents on Acanthamoeba sp. and evidence of a natural resilience to amphotericin B. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, X.; Wei, P.; Su, X.; Zhao, L.; Wu, M.; Hao, C.; Liu, C.; Zhao, D.; Cheng, M. Design, synthesis and evaluation of aromatic heterocyclic derivatives as potent antifungal agents. Eur. J. Med. Chem. 2017, 137, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wei, P.; Wu, M.; Zhang, X.; Zhao, L.; Jiang, X.; Hao, C.; Su, X.; Zhao, D.; Cheng, M. Design, synthesis and evaluation of benzoheterocycle analogues as potent antifungal agents targeting CYP51. Bioorg. Med. Chem. 2018, 26, 3242–3253. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Neugebauer, T.; Zill, P.; Lass-Florl, C.; Bracher, F.; Binder, U. Sterol Composition of Clinically Relevant Mucorales and Changes Resulting from Posaconazole Treatment. Molecules 2018, 23, 1218. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shi, H.; Chang, W.; Li, Y.; Zhang, M.; Qiao, Y.; Lou, H. Eudesmane sesquiterpenes from Chinese liverwort are substrates of Cdrs and display antifungal activity by targeting Erg6 and Erg11 of Candida albicans. Bioorg. Med. Chem. 2017, 25, 5764–5771. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, S.B.; Karkare, S.; Bergdoll, M.; Rahier, A.; Leighton-Davis, J.R.; Fioretto, C.; Aust, T.; Filipuzzi, I.; Frederiksen, M.; Gounarides, J.; et al. FR171456 is a specific inhibitor of mammalian NSDHL and yeast Erg26p. Nat. Commun. 2015, 6, 8613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warrilow, A.G.; Parker, J.E.; Price, C.L.; Nes, W.D.; Garvey, E.P.; Hoekstra, W.J.; Schotzinger, R.J.; Kelly, D.E.; Kelly, S.L. The Investigational Drug VT-1129 Is a Highly Potent Inhibitor of Cryptococcus Species CYP51 but Only Weakly Inhibits the Human Enzyme. Antimicrob. Agents Chemother. 2016, 60, 4530–4538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhanout, K.; Rolain, J.M.; Brunel, J.M. Squalamine as an example of a new potent antimicrobial agents class: A critical review. Curr. Med. Chem. 2010, 17, 3909–3917. [Google Scholar] [CrossRef] [PubMed]
- Savage, P.B.; Li, C.; Taotafa, U.; Ding, B.; Guan, Q. Antibacterial properties of cationic steroid antibiotics. FEMS Microbiol. Lett. 2002, 217, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Tseng, W.R.; Ahmed, A.F.; Chiang, P.L.; Tai, C.J.; Hwang, T.L.; Dai, C.F.; Sheu, J.H. Anti-Inflammatory Polyoxygenated Steroids from the Soft Coral Lobophytum michaelae. Mar. Drugs 2018, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.M.; Ito, T.; Win, N.N.; Vo, H.Q.; Nguyen, H.T.; Morita, H. A new sterol from the Vietnamese marine sponge Xestospongia testudinaria and its biological activities. Nat. Prod. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Sekine, A.; Fukazawa, H.; Uehara, Y.; Yamaguchi, K.; Endo, Y.; Okuda, T.; Furumai, T.; Oki, T. Anicequol, a novel inhibitor for anchorage-independent growth of tumor cells from Penicillium aurantiogriseum Dierckx TP-F0213. J. Antibiot. 2002, 55, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, Y.; Sakai, N.; Matsumoto, K.; Mizoue, K. A novel neuritogenic compound, NGA0187. J. Antibiot. 2002, 55, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.R.; Noguchi, K.; Fukazawa, H.; Igarashi, Y.; Arai, H.; Uehara, Y. p38MAPK and Rho-dependent kinase are involved in anoikis induced by anicequol or 25-hydroxycholesterol in DLD-1 colon cancer cells. Biochem. Biophys. Res. Commun. 2013, 430, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.M.; Liu, S.X.; Huang, C.H.; Pang, J.Y.; Lin, Y.C. Secondary metabolites of a mangrove endophytic fungus Aspergillus terreus (No. GX7-3B) from the South China Sea. Mar. Drugs 2013, 11, 2616–2624. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.S.; Li, X.M.; Li, C.S.; Proksch, P.; Wang, B.G. Penicisteroids A and B, antifungal and cytotoxic polyoxygenated steroids from the marine alga-derived endophytic fungus Penicillium chrysogenum QEN-24S. Bioorg. Med. Chem. Lett. 2011, 21, 2894–2897. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razek, A.S.; Hamed, A.; Frese, M.; Sewald, N.; Shaaban, M. Penicisteroid C: New polyoxygenated steroid produced by co-culturing of Streptomyces piomogenus with Aspergillus niger. Steroids 2018, 138, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.B.; Xiao, H.; Fan, C.Q.; Lu, Y.N.; Zhang, G.; Yin, S. Bioactive polyhydroxylated sterols from the marine sponge Haliclona crassiloba. Steroids 2013, 78, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, N.T.; Huong, P.T.; Thanh, N.V.; Chi, N.T.; Dang, N.H.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; Kiem, P.V.; Minh, C.V. Cytotoxic Steroids from the Vietnamese Soft Coral Sinularia conferta. Chem. Pharm. Bull. 2017, 65, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, H.; Wang, P.; Gong, W.; Xue, M.; Zhang, H.; Liu, T.; Liu, B.; Yi, Y.; Zhang, W. Bioactive polyoxygenated steroids from the South China sea soft coral, Sarcophyton sp. Mar. Drugs 2013, 11, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.W.; Su, J.H.; Lin, C.C.; Li, Y.R.; Chao, Y.H.; Lin, S.H.; Chan, H.L. 24-Methyl-Cholesta-5,24(28)-Diene-3beta,19-diol-7beta-Monoacetate Inhibits Human Small Cell Lung Cancer Growth In Vitro and In Vivo via Apoptosis Induction. Mar. Drugs 2017, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Meesala, S.; Gurung, P.; Karmodiya, K.; Subrayan, P.; Watve, M.G. Isolation and structure elucidation of halymeniaol, a new antimalarial sterol derivative from the red alga Halymenia floresii. J. Asian Nat. Prod. Res. 2018, 20, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, X.C.; Longeon, A.; Pham, V.C.; Urvois, F.; Bressy, C.; Trinh, T.T.; Nguyen, H.N.; Phan, V.K.; Chau, V.M.; Briand, J.F.; et al. Antifouling 26,27-cyclosterols from the Vietnamese marine sponge Xestospongia testudinaria. J. Nat. Prod. 2013, 76, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Pailee, P.; Mahidol, C.; Ruchirawat, S.; Prachyawarakorn, V. Sterols from Thai Marine Sponge Petrosia (Strongylophora) sp. and Their Cytotoxicity. Mar. Drugs 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Liu, D.; de Voogd, N.J.; Proksch, P.; Lin, W. New sterol derivatives from the marine sponge Xestospongia sp. Helvetica. Chim. Acta 2016, 99, 588–596. [Google Scholar] [CrossRef]
- Tsai, C.-R.; Huang, C.Y.; Chen, B.W.; Tsai, Y.Y.; Shih, S.-P.; Hwang, T.-L.; Dai, C.-F.; Wang, S.-Y.; Sheu, J.-H. New bioactive steroids from the soft coral Klyxum flaccidum. RSC Adv. 2015, 5, 12546–12554. [Google Scholar] [CrossRef]
- Tseng, W.R.; Huang, C.Y.; Tsai, Y.Y.; Lin, Y.S.; Hwang, T.L.; Su, J.H.; Sung, P.J.; Dai, C.F.; Sheu, J.H. New cytotoxic and anti-inflammatory steroids from the soft coral Klyxum flaccidum. Bioorg. Med. Chem. Lett. 2016, 26, 3253–3257. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, F.; Kudo, N.; Tomachi, Y.; Nakata, A.; Takemoto, M.; Ito, A.; Tabei, H.; Arai, D.; de Voogd, N.; Yoshida, M.; et al. Halistanol sulfates I. and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria. J. Antibiot. 2018, 71, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Distrutti, E.; Bifulco, G.; D’Auria, M.V.; Zampella, A. Marine sponge steroids as nuclear receptor ligands. Trends Pharmacol. Sci. 2012, 33, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, H.; Sun, X.; Zhao, H.; Wu, L.; Zhu, D.; Yang, G.; Shao, Y.; Zhang, X.; Mao, X.; et al. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Metwaly, A.M.; Kadry, H.A.; El-Hela, A.A.; Mohammad, A.I.; Ma, G.; Cutler, S.J.; Ross, S.A. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem. Lett. 2014, 7, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Wang, K.L.; Liu, M.; She, Z.G.; Wang, C.Y. Bioactive steroid derivatives and butyrolactone derivatives from a gorgonian-derived Aspergillus sp. fungus. Chem. Biodivers. 2015, 12, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, G.D.; Feng, X.L.; Yu, Y.; He, R.R.; Li, X.X.; Huang, Y.; Zhou, W.X.; Guo, L.D.; Zheng, Y.Z.; et al. Nodulisporiviridins A-H, Bioactive Viridins from Nodulisporium sp. J. Nat. Prod. 2015, 78, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Kehraus, S.; Nesaei-Mosaferan, D.; Hufendieck, P.; Meijer, L.; Konig, G.M. Abeta-42 lowering agents from the marine-derived fungus Dichotomomyces cejpii. Steroids 2015, 104, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, N.T.; Huong, P.T.; Thanh, N.V.; Cuong, N.X.; Nam, N.H.; Thung do, C.; Kiem, P.V.; Minh, C.V. Steroid Constituents from the Soft Coral Sinularia nanolobata. Chem. Pharm. Bull. 2016, 64, 1417–1419. [Google Scholar] [CrossRef] [PubMed]
- Thanh, N.V.; Ngoc, N.T.; Anh Hle, T.; Thung do, C.; Thao do, T.; Cuong, N.X.; Nam, N.H.; Kiem, P.V.; Minh, C.V. Steroid constituents from the soft coral Sinularia microspiculata. J. Asian Nat. Prod. Res. 2016, 18, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, N.T.; Hanh, T.T.H.; Thanh, N.V.; Thao, D.T.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; Kiem, P.V.; Minh, C.V. Cytotoxic Steroids from the Vietnamese Soft Coral Sinularia leptoclados. Chem. Pharm. Bull. 2017, 65, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.H.T.; Nguyen Viet, P.; Nguyen Van, T.; Tran, H.T.; Nguyen Xuan, C.; Nguyen Hoai, N.; Do Cong, T.; Phan Van, K.; Chau Van, M. Cytotoxic steroid derivatives from the Vietnamese soft coral Sinularia brassica. J. Asian Nat. Prod. Res. 2017, 19, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Chen, W.T.; Yao, L.G.; Guo, Y.W. Two new cytotoxic steroids from the Chinese soft coral Sinularia sp. Steroids 2018, 136, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Su, J.H.; Liaw, C.C.; Sung, P.J.; Chiang, P.L.; Hwang, T.L.; Dai, C.F.; Sheu, J.H. Bioactive Steroids with Methyl Ester Group in the Side Chain from a Reef Soft Coral Sinularia brassica Cultured in a Tank. Mar. Drugs 2017, 15, 280. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.Y.; Huang, C.Y.; Tseng, W.R.; Chiang, P.L.; Hwang, T.L.; Su, J.H.; Sung, P.J.; Dai, C.F.; Sheu, J.H. Klyflaccisteroids, K-M bioactive steroidal derivatives from a soft coral Klyxum flaccidum. Bioorg. Med. Chem. Lett. 2017, 27, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Ren, J.; Huang, Q.; Long, H.; Jin, H.; Zhang, L.; Liu, H.; van Ofwegen, L.; Lin, W. Pregnane steroids from a gorgonian coral Subergorgia suberosa with anti-flu virus effects. Steroids 2016, 108, 99–104. [Google Scholar] [CrossRef] [PubMed]
- He, W.F.; Xue, D.Q.; Yao, L.G.; Li, J.; Liu, H.L.; Guo, Y.W. A new bioactive steroidal ketone from the South China Sea sponge Xestospongia testudinaria. J. Asian Nat. Prod. Res. 2016, 18, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Mencarelli, A.; D’Amore, C.; Renga, B.; Cipriani, S.; Carino, A.; Sepe, V.; Perissutti, E.; D’Auria, M.V.; Zampella, A.; Distrutti, E.; et al. Solomonsterol A, a marine pregnane-X-receptor agonist, attenuates inflammation and immune dysfunction in a mouse model of arthritis. Mar. Drugs 2013, 12, 36–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, Y.Y.; Tang, J.; Sun, F.; Lin, H.W. New 4-methylidene sterols from the marine sponge Theonella swinhoei. Fitoterapia 2018, 127, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Cheng, C.; Reimer, A.; Kozjak-Pavlovic, V.; Ibrahim, A.K.; Rudel, T.; Hentschel, U.; Edrada-Ebel, R.; Ahmed, S.A. Antichlamydial sterol from the Red Sea sponge Callyspongia aff. implexa. Planta. Med. 2015, 81, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Perumal, P.; Sowmiya, R.; Prasanna Kumar, S.; Ravikumar, S.; Deepak, P.; Balasubramani, G. Isolation, structural elucidation and antiplasmodial activity of fucosterol compound from brown seaweed, Sargassum linearifolium against malarial parasite Plasmodium falciparum. Nat. Prod. Res. 2018, 32, 1316–1319. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Cho, Y.R.; Hong, S.S.; Anh, E.K. Anti-obesity activity of saringosterol isolated from Sargassum muticum (Yendo) fensholt extract in 3t3-L1 cells. Phytother. Res. 2017, 31, 1694–1701. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.G.; Zhou, M.; Jin, Q.H.; Liu, B.; Guan, L.P. Antidepressant-like effects of saringosterol, a sterol from Sargassum fusiforme by performing in vivo behavioral tests. Med. Chem. Res. 2017, 26, 909–915. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, L.W.; Feng, M.T.; Liu, A.H.; Li, J.; Zhao, T.S.; Lai, X.P.; Wang, B.; Guo, Y.W.; Mao, S.C. Dictyoptesterols A–C, ∆22-24-oxo cholestane-type sterols with potent PTP1B inhibitory activity from the brown alga Dictyopteris undulata Holmes. Fitoterapia 2018, 130, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.T.; Wang, T.; Liu, A.H.; Li, J.; Yao, L.G.; Wang, B.; Guo, Y.W.; Mao, S.C. PTP1B inhibitory and cytotoxic C-24 epimers of Delta(28)-24-hydroxy stigmastane-type steroids from the brown alga Dictyopteris undulata Holmes. Phytochemistry 2018, 146, 25–35. [Google Scholar] [CrossRef] [PubMed]









| Algal Organism | Major Sterols 1 (>40%) | Semi-Major Sterols 1 (>20%) | Minor Sterols 1 (<20%) | Reference 2 |
|---|---|---|---|---|
| Ulvophyceae | ||||
| Acetabularia caliculus | (3/37) | (36/38), (39/40), (41/42), 44, (48/49) | [52] | |
| Acicularia schenckii | (3/37) | (36/38), (39/40), (41/42), 44 | [52] | |
| Trebouxiophyceae | ||||
| Chlorella vulgaris | 2 | 52 | 56, 58, 64, 62, 66 | [53] |
| Chlorella luteoviridis | 40 | 38 | 37, 42, 52 | [53] |
| Eustigmatophyceae | ||||
| Nannochloropsis limnetica | 1 | 44, (3/37), 45, (67/68) | [53] | |
| Bacillariophyceae (diatoms) | ||||
| Stephanodiscus hantzschii | 45 | 48, 17, 57, 46 | [53] | |
| Gomphonema parvulum | 41 | (69/2), (59/60), (36,38) | [53] | |
| Cyclotella meneghiniana | 45 | 38, 43, 48 | [53] | |
| Raphidophyceae | ||||
| Chloromorum toxicum | 40 | 37, 1, 42, 38, 48 | [54] | |
| Chattonella marina | 3 | 1, 63 | [54] | |
| Heterosigma akashiwo | 37 | 38 | [54] | |
| Dictyochophyceae | ||||
| Verrucophora farcimen | 70 | [54] | ||
| Chlorophyceae (see also Figure 3) | ||||
| Scenedesmus obliquus | 54 | 52 | 50, 62, 60 | [53] |
| Monoraphidium mintutum | 50 | 52 | 65, 54, 62, 60, 64 | [53] |
| Cryptophyceae | ||||
| Cryptomonas sp. | 39, 41 | [53] | ||
| Rhodomonas sp. | 41 | [55] | ||
| No.1 | Microbial Source | Biological Target 2 | Biological Activity | Reference |
|---|---|---|---|---|
| Fungi | ||||
| 163 | Nigrospora sphaerica | Cryptococcus neoformans | IC50 14.81 µg/mL | [106] |
| 164 | Gymnoascus reessii | NCI-H187 | IC50 16.3 µg/mL | [34] |
| Plasmodium falciparum | IC50 3.3 µg/mL | [34] | ||
| 165 | Gymnoascus reessii | NCI-H187 | IC50 47.9 µg/mL | [34] |
| Plasmodium falciparum | IC50 4.5 µg/mL | [34] | ||
| 166 | Gymnoascus reessii | NCI-H187 | IC50 1.9 µg/mL | [34] |
| Plasmodium falciparum | IC50 3.4 µg/mL | [34] | ||
| 167 | Gymnoascus reessii | NCI-H187 | IC50 12.5 µg/mL | [34] |
| Plasmodium falciparum | IC50 3.4 µg/mL | [34] | ||
| 168 | Aspergillus sp. | Balanus amphitrite biofouling | EC50 18.40 µg/mL | [107] |
| 169 | Nodulisporium sp. | Aβ42 aggregation | IC50 10.1 µM | [108] |
| 170 | Nodulisporium sp. | Aβ42 aggregation | 54.6% relative inhibitory activity at 100 µM | [108] |
| 171 | Nodulisporium sp. | Aβ42 aggregation | IC50 1.2 µM | [108] |
| 172 | Nodulisporium sp. | Aβ42 aggregation | IC50 43.5 µM | [108] |
| 173 | Dichotomomyces cejpii | Aβ42 aggregation | pretreatment with 10 µM reduced production of Aβ peptides to 3.8-fold increase with 100 µM Aftin-5 compared to 9.4-fold increase with only Aftin-5 and no inhibitor | [109] |
| Coral | ||||
| 174 | Sinularia nanolobata | HL-60 | IC50 33.53 µM | [110] |
| HepG2 | IC50 64.35 µM | [110] | ||
| 175 | Sinularia microspiculata | HL-60 | IC50 82.80 µM | [111] |
| SK-Mel2 | IC50 72.32 µM | [111] | ||
| 176 | Sinularia leptoclados | HL-60 | IC50 13.45 µM | [112] |
| SW480 | IC50 14.42 µM | [112] | ||
| LNCaP | IC50 17.13 µM | [112] | ||
| MCF-7 | IC50 17.29 µM | [112] | ||
| 177 | Sinularia leptoclados | HL-60 | IC50 20.53 µM | [112] |
| SW480 | IC50 26.61 µM | [112] | ||
| KB | IC50 32.86 µM | [112] | ||
| 178 | Sinularia conferta | A549 | IC50 78.73 µM | [94] |
| HeLa | IC50 30.5 µM | [94] | ||
| PANC-1 | IC50 9.35 µM | [94] | ||
| 179 | Sinularia conferta | A549 | IC50 27.12 µM | [94] |
| HeLa | IC50 24.64 µM | [94] | ||
| PANC-1 | IC50 20.51 µM | [94] | ||
| 180 | Sinularia brassica | PANC-1 | IC50 15.24 µM | [113] |
| A549 | IC50 39.36 µM | [113] | ||
| 181 | Sinularia brassica | PANC-1 | IC50 22.47 µM | [113] |
| A549 | IC50 41.20 µM | [113] | ||
| 182 | Sinularia brassica | A549 | IC50 47.31 µM | [113] |
| 183 | Sinularia brassica | PANC-1 | IC50 15.39 µM | [113] |
| A549 | IC50 47.46 µM | [113] | ||
| 184 | Sinularia brassica | PANC-1 | IC50 38.12 µM | [113] |
| A549 | IC50 23.73 µM | [113] | ||
| 185 | Sinularia brassica | A549 | IC50 92.53 µM | [113] |
| Sacrcophyton sp. | E. coli | 0.05 mg ZOI 3 10.0 mm | [95] | |
| S. tritici | 0.05 mg ZOI 7.5 mm | [95] | ||
| 187 | Sinularia microspiculata | HL-60 | IC50 89.02 µM | [111] |
| 188 | Sinularia sp. | HL-60 | IC50 1.79 µM | [114] |
| 189 | Sinularia sp. | HL-60 | IC50 4.03 µM | [114] |
| 190 | Sinularia sp. | HL-60 | IC50 0.69 µM | [114] |
| 191 | Sinularia brassica | P388D1 | IC50 37.2 µM | [115] |
| MOLT-4 | IC50 37.8 µM | [115] | ||
| 192 | Sinularia brassica | P388D1 | IC50 9.7 µM | [115] |
| MOLT-4 | IC50 6.0 µM | [115] | ||
| 193 | Sinularia brassica | P388D1 | IC50 5.7 µM | [115] |
| MOLT-4 | IC50 5.3 µM | [115] | ||
| 194 | Sinularia brassica | P388D1 | IC50 24.4 µM | [115] |
| MOLT-4 | IC50 31.2 µM | [115] | ||
| 186 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 5.0 mm | [95] |
| S. tritici | 0.05 mg ZOI 7.0 mm | [95] | ||
| 195 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 7.5 mm | [95] |
| S. tritici | 0.05 mg ZOI 10.5 mm | [95] | ||
| 196 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 4.5 mm | [95] |
| S. tritici | 0.05 mg ZOI 6.5 mm | [95] | ||
| 197 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 6.0 mm | [95] |
| S. tritici | 0.05 mg ZOI 4.5 mm | [95] | ||
| 198 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 6.0 mm | [95] |
| S. tritici | 0.05 mg ZOI 6.0 mm | [95] | ||
| 199 | Sacrcophyton sp. | E. coli | 0.05 mg ZOI 6.0 mm | [95] |
| S. tritici | 0.05 mg ZOI 9.0 mm | [95] | ||
| 200 | Klyxum flaccidum | A549 | ED50 7.7 µg/mL | [101] |
| 201 | Klyxum flaccidum | K562 | IC50 12.7 µM | [102] |
| elastase release | IC50 4.40 µM | [102] | ||
| 202 | Klyxum flaccidum | P388 | IC50 31.8 µM | [116] |
| elastase release | IC50 5.84 µM | [116] | ||
| 203 | Subergorgia suberosa | Influenza virus strain A/WSN/33 (H1N1) | IC50 37.73 µM | [117] |
| 204 | Subergorgia suberosa | Influenza virus strain A/WSN/33 (H1N1) | IC50 50.95 µM | [117] |
| Sponges | ||||
| 205 | Petrosia sp. | MOLT-3 | IC50 36.57 µM | [99] |
| A549 | IC50 54.26 µM | [99] | ||
| 206 | Xestospongia testudinaria | MCF-7 | IC50 55.8 µM | [86] |
| A549 | IC50 63.1 µM | [86] | ||
| 207 | Xestospongia testudinaria | PTP1B 4 | IC50 4.27 µM | [118] |
| 208 | Xestospongia sp. | K562 | IC50 18.32 µM | [100] |
| 209 | Xestospongia sp. | K562 | 25.73% inhibition at 10 µM | [100] |
| 210 | Xestospongia sp. | K562 | 41.32% inhibition at 10 µM | [100] |
| 158 | Theonella swinhoei | arthritis | 30% reduction in clinical arthritis scores in mice treated with 10 mg/kg | [119] |
| 211 | Theonella swinhoei | U937 | IC50 8.8 µM | [120] |
| PC-9 | IC50 7.7 µM | [120] | ||
| 212 | Theonella swinhoei | U937 | IC50 2.0 µM | [120] |
| PC-9 | IC50 9.7 µM | [120] | ||
| 213 | Theonella swinhoei | U937 | IC50 3.2 µM | [120] |
| PC-9 | IC50 1.6 µM | [120] | ||
| 214 | Callyspongia aff. implexa | Chlamydia trachomatis | IC50 2.3 µM | [121] |
| Brown Algae | ||||
| 44 | Sargassum linearifolium | Plasmodium falciparum | IC50 7.48 µg/mL | [122] |
| 215 | Sargassum muticum | obesity | decreased lipid accumulation and dose-dependent suppression of PPARγ 5 | [123] |
| Sargassum fusiform | depression | 32.67/53.60 and 32.06/50.83 percentage decrease in immobility duration for forced swimmin and tail suspension test in the mouse model at 10 mg/kg/30 mg/kg | [124] | |
| 216 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 7.92 µM | [125] |
| 217 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 7.78 µM | [125] |
| 218 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 3.03 µM | [125] |
| 219 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 3.72 µM | [125] |
| 220 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 15.01µM | [125] |
| 221 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 35.01 µM | [126] |
| 222 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 1.88 µM | [126] |
| HL-60 | IC50 2.08 µM | [126] | ||
| 223 | Dictyopteris undulata Holmes | HL-60 | IC50 2.45 µM | [126] |
| 224 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 38.15 µM | [126] |
| HL-60 | IC50 2.70 µM | [126] | ||
| 225 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 48.21 µM | [126] |
| HL-60 | IC50 1.02 µM | [126] | ||
| 226 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 3.47 µM | [126] |
| HL-60 | IC50 1.26 µM | [126] | ||
| 227 | Dictyopteris undulata Holmes | PTP1B 4 | IC50 16.03 µM | [126] |
| HL-60 | IC50 1.17 µM | [126] | ||
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haubrich, B.A. Microbial Sterolomics as a Chemical Biology Tool. Molecules 2018, 23, 2768. https://doi.org/10.3390/molecules23112768
Haubrich BA. Microbial Sterolomics as a Chemical Biology Tool. Molecules. 2018; 23(11):2768. https://doi.org/10.3390/molecules23112768
Chicago/Turabian StyleHaubrich, Brad A. 2018. "Microbial Sterolomics as a Chemical Biology Tool" Molecules 23, no. 11: 2768. https://doi.org/10.3390/molecules23112768