Identification of Cyclic Dipeptides from Escherichia coli as New Antimicrobial Agents against Ralstonia Solanacearum
Abstract
:1. Introduction
2. Results
2.1. Isolation of New Antagonistic Bacteria against R. solanacearum
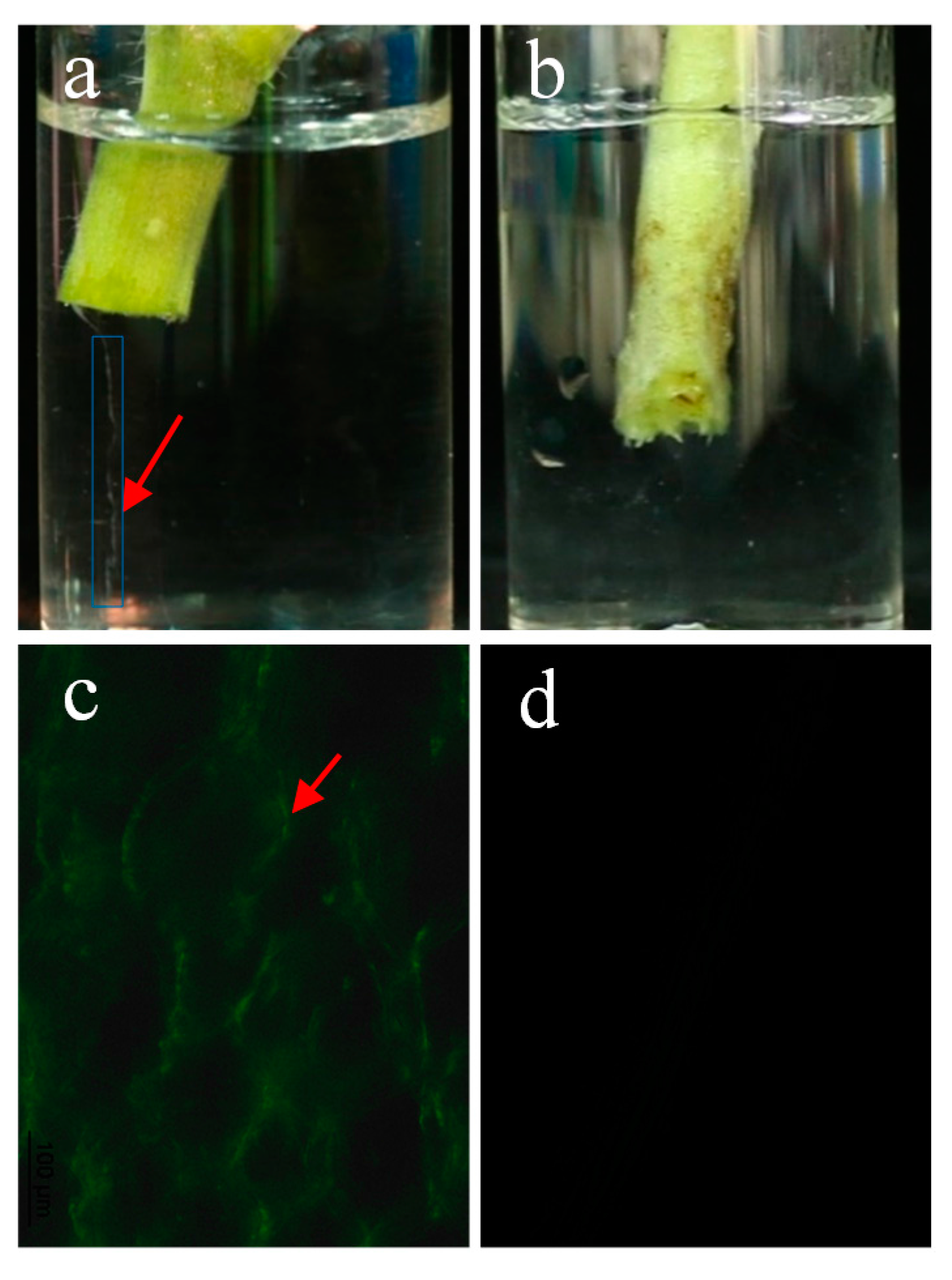
2.2. E. coli GZ-34 Shows Effective Protection to Tomato from R. solanacearum Infection
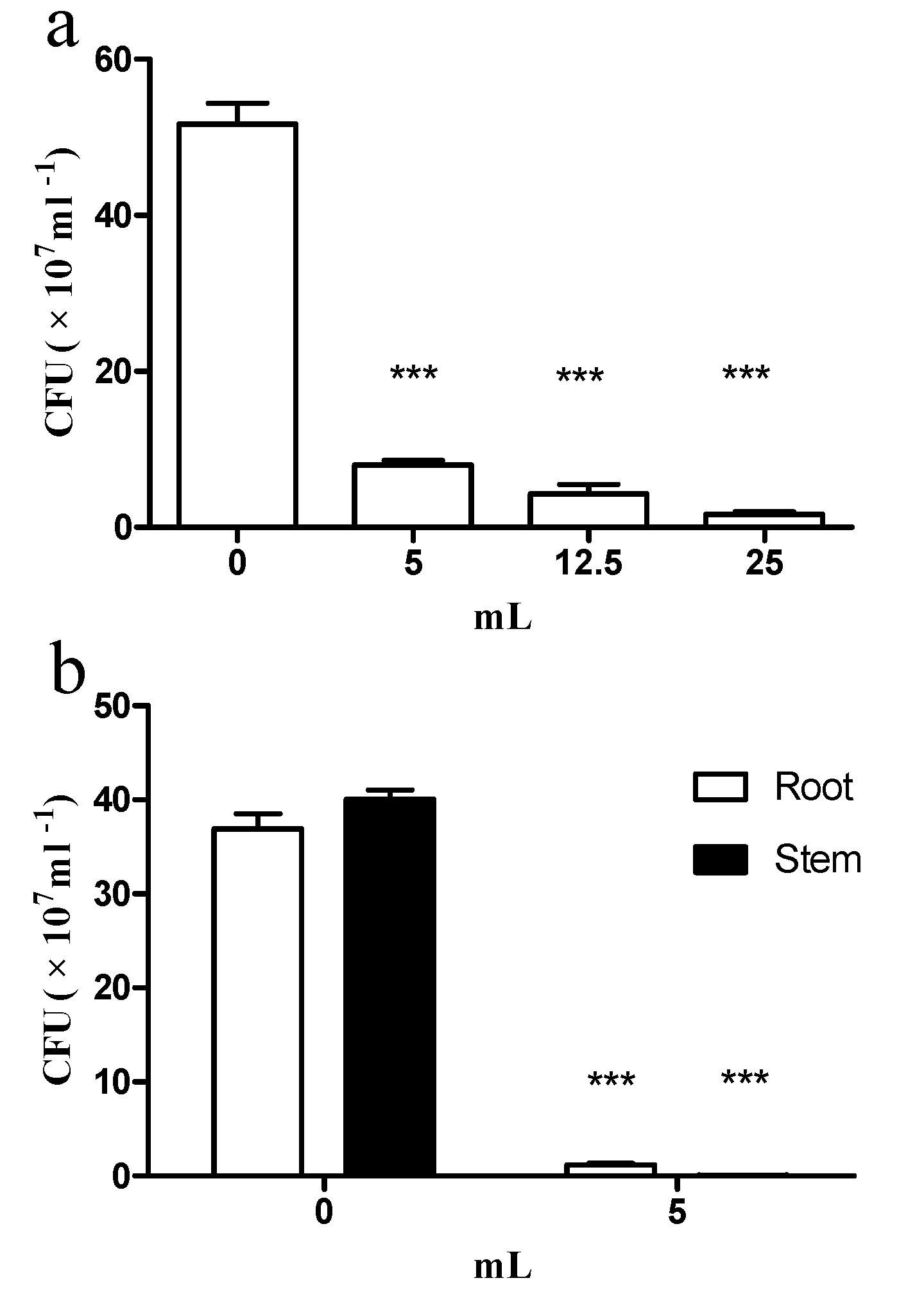
2.3. E. coli GZ-34 Remarkably Inhibits the Cell Growth of R. solanacearum in Soil and Plants
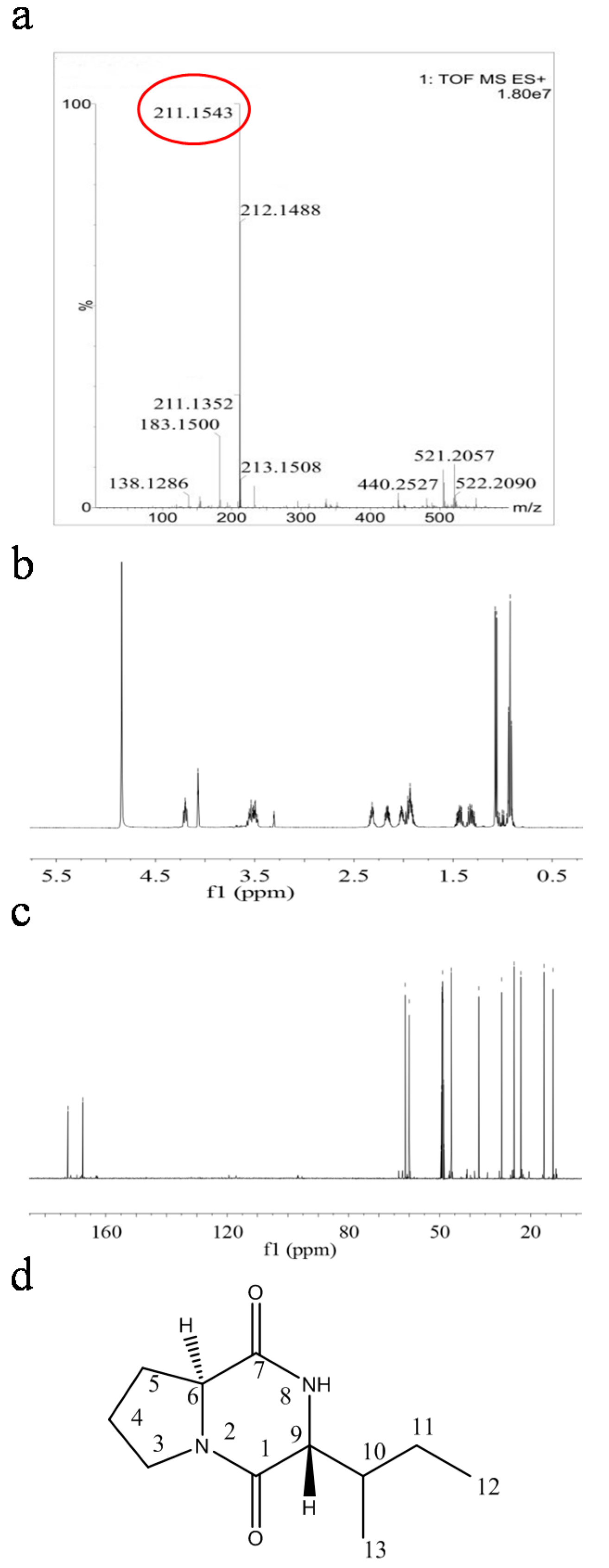
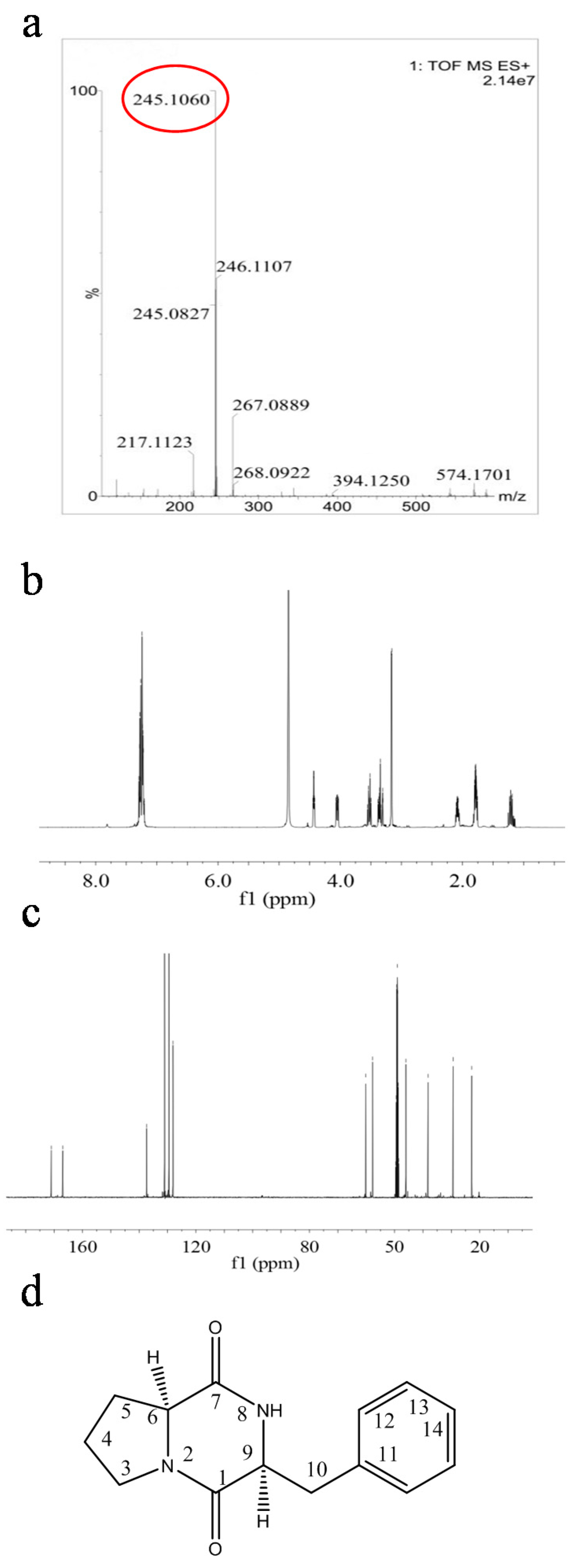
2.4. Structural Characterization of Antimicrobial Compounds Isolated from E. coli GZ-34
2.5. Antimicrobial Compounds Isolated from E. coli GZ-34 Interfere with Cell Growth and Expression Levels of Virulence Contributors of R. solanacearum
2.6. Cyclo(l-Pro-l-Phe) Inhibits Spore Formation in M. Grisea
3. Discussion
4. Materials and Methods
4.1. Bacterial and Fungal Growth Conditions and MIC Assays
4.2. Isolation of Antagonistic Bacteria against R. solanacearum
4.3. Characterization of Antagonistic Bacteria against R. solanacearum
4.4. Construction of GMI1000(eGFP) Strain
4.5. Analysis of Efficacy against Tomato Bacterial Wilt
4.6. Analysis of R. solanacearum Cell Numbers in Soil and in Plants
4.7. Analysis of Antimicrobial Compounds
4.8. Microscopic Analysis and Quantification of Spore Formation in M. grisea
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kleman, A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 1954, 44, 683–685. [Google Scholar]
- Wicker, E.; Grassart, L.; Coransonbeaudu, R.; Mian, D.; Guilbaud, C.; Fegan, M.; Prior, P. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 2007, 73, 6790–6801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, N.; Guidot, A.; Vailleau, F.; Valls, M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013, 14, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, C.A.; Rossato, M.; Boiteux, L.S. The host status of coffee (coffea arabica) to Ralstonia solanacearum phylotype I isolates. Trop. Plant Pathol. 2015, 40, 1–4. [Google Scholar] [CrossRef]
- Goto, M. Fundamentals of bacterial plant pathology; Academic Press, Inc.: San Diego, CA, USA, 1992; pp. 110–155. ISBN 9780323140447. [Google Scholar]
- Prior, P.; Ailloud, F.; Dalsing, B.L.; Remenant, B.; Sanchez, B.; Allen, C. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genom. 2016, 17, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravelomanantsoa, S.; Robène, I.; Chiroleu, F.; Guérin, F.; Poussier, S.; Pruvost, O.; Prior, P. A novel multilocus variable number tandem repeat analysis typing scheme for african phylotype III strains of the Ralstonia solanacearum species complex. PeerJ 2016, 4, 1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ailloud, F.; Lowe, T.M.; Robène, I.; Cruveiller, S.; Allen, C.; Prior, P. In planta comparative transcriptomics of host-adapted strains of Ralstonia solanacearum. PeerJ 2016, 4, 1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Yan, L.; Lei, Y.; Jiang, H.; Ren, X.; Liao, B. Expressed sequence tags in cultivated peanut (arachis hypogaea): Discovery of genes in seed development and response to Ralstonia solanacearum challenge. J. Plant Res. 2012, 125, 755. [Google Scholar] [CrossRef] [PubMed]
- Perrier, A.; Peyraud, R.; Rengel, D.; Barlet, X.; Lucasson, E.; Gouzy, J.; Peeters, N.; Genin, S.; Guidot, A. Enhancedin plantafitness through adaptive mutations in efpr, a dual regulator of virulence and metabolic functions in the plant pathogen Ralstonia solanacearum. PLoS Pathog. 2016, 12, e1006044. [Google Scholar] [CrossRef] [PubMed]
- Kazuhiro, N.; Caitilyn, A. A pectinase-deficient Ralstonia solanacearum strain induces reduced and delayed structural defences in tomato xylem. J. Phytopathol. 2009, 157, 228–234. [Google Scholar] [CrossRef]
- Hayward, A.C. Biology and epidemiology of bacterial wilt cause by Pseudomona solanacearum. Annu. Rev. Phytopathol. 1991, 29, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.C. Ralstonia solanacearum. In Encyclopedia of Microbiology; Lederberg, J., Ed.; Academic Press: San Diego, CA, USA, 2000; Volume 4, pp. 32–42. ISBN 9780122268021. [Google Scholar]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microbiol. 2011, 77, 4155. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.; Staskawicz, B. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365. [Google Scholar] [CrossRef] [PubMed]
- Wagner, W.C. Sustainable agriculture: How to sustain a production system in a changing environment. Int. J. Parasitol. 1999, 29, 1–5. [Google Scholar] [CrossRef]
- Pierre, U. Sustainable agriculture and chemical control: Opponents or components of the same strategy. Crop. Prot. 2000, 19, 831–836. [Google Scholar] [CrossRef]
- Chave, M.; Crozilhac, P.; Deberdt, P.; Plouznikoff, K.; Declerck, S. Rhizophagus irregularis MUCL 41833 transitorily reduces tomato bacterial wilt incidence caused by Ralstonia solanacearum, under in vitro conditions. Mycorrhiza 2017, 27, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Raza, W.; Ling, N.; Liu, D.; Wei, Z.; Huang, Q.; Shen, Q. Volatile organic compounds produced by Pseudomonas fluorescens WR-1 restrict the growth and virulence traits of Ralstonia solanacearum. Microbiol. Res. 2016, 192, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Raza, W.; Wei, Z.; Ling, N.; Huang, Q.; Shen, Q. Effect of organic fertilizers prepared from organic waste materials on the production of antibacterial volatile organic compounds by two biocontrol Bacillus amyloliquefaciens strains. J. Biotechnol. 2016, 227, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Elhalag, K.M.; Messiha, N.A.; Emara, H.M.; Abdallah, S.A. Evaluation of antibacterial activity of Stenotrophomonas maltophilia against Ralstonia solanacearum under different application conditions. J. Appl. Microbiol. 2016, 6, 1629–1645. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.N.; Sreekala, S.R.; Chandrasekaran, D.; Nambisan, B.; Anto, R.J. Biocontrol of aspergillus species on peanut kernels by antifungal diketopiperazine producing Bacillus cereus associated with entomopathogenic nematode. PLoS ONE 2014, 9, e106041. [Google Scholar] [CrossRef] [PubMed]
- Nishanth, K.S.; Mohandas, C.; Siji, J.V.; Rajasekharan, K.N.; Nambisan, B. Identification of antimicrobial compound, diketopiperazines, from a Bacillus sp. N strain associated with a rhabditid entomopathogenic nematode against major plant pathogenic fungi. J. Appl. Microbiol. 2012, 113, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Musetti, R.; Polizzotto, R.; Vecchione, A.; Borselli, S.; Zulini, L.; D’Ambrosio, M.; di Toppi, L.S.; Pertot, I. Antifungal activity of diketopiperazines extracted from Alternaria alternata against Plasmopara viticola: An ultrastructural study. Micron 2007, 38, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Dong, J.; Zhou, X.; Yang, X.; Lee, K.; Wang, L.; Zhang, S.; Liu, Y. Proline-containing dipeptides from a marine sponge of a Callyspongia species. Helvetica Chim. Acta 2010, 92, 1112–1117. [Google Scholar] [CrossRef]
- Furtado, N.A.; Pupo, M.T.; Carvalho, I.; Campo, V.L.; Duarte, M.C.T.; Bastos, J.K. Diketopiperazines produced by an Aspergillus fumigatus brazilian strain. J. Braz. Chem. Soc. 2005, 16, 1448–1453. [Google Scholar] [CrossRef]
- Giessen, T.; Vontesmar, A.; Marahiel, M. Insights into the generation of structural diversity in a tRNA-dependent pathway for highly modified bioactive cyclic dipeptides. Chem. Biol. 2013, 20, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.J.J.; Weger, L.A.D.; Bennett, J.W. Microbial stimulation of plant growth and protection from disease. Curr. Opin. Biotechnol. 1991, 2, 457–464. [Google Scholar] [CrossRef]
- Yan, M.; Zhu, G.; Lin, S.; Xian, X.; Chang, C.; Xi, P.; Shen, W.; Huang, W.; Cai, E.; Jiang, Z. The Mating-type Locus b of the Sugarcane Smut Sporisorium scitamineum is essential for mating, filamentous growth and pathogenicity. Fungal Genet. Biol. 2016, 86, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Prasad, C. Bioactive cyclic dipeptides. Peptides 1995, 16, 151–164. [Google Scholar] [CrossRef]
- Dinsmore, C.J.; Beshore, D.C. Recent advances in the synthesis of diketopiperazines. Tetrahedron 2002, 58, 3297–3312. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, X.; Xu, T.; Yang, X.; Liu, Y. Diketopiperazines from marine organisms. Chem. Biodivers. 2010, 7, 2809. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Yan, T.; Peng, Y.; Zhou, X.; Yang, X.; Liu, Y. Diketopiperazines from the marine sponge Axinella sp. Chem. Nat. Compd. 2014, 50, 191–193. [Google Scholar] [CrossRef]
- Campbell, J.; Lin, Q.; Geske, G.D.; Blackwell, H.E. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 2009, 4, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Sauguet, L.; Moutiez, M.; Li, Y.; Belin, P.; Seguin, J.; Du, M.L.; Thai, R.; Masson, C.; Fonvielle, M.; Pernodet, J.L. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 2011, 39, 4475–4489. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chem. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- King, B.D.; Elder, J.D.; Proctor, D.F.; Dripps, R.D. Reflex circulatory responses to tracheal intubation performed under topical anesthesia. Anesthesiology 1954, 15, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Schaad, A.; Moffett, J.; Jacob, J. The role-based access control system of a European bank: A case study and discussion. In Proceedings of the sixth ACM Symposium on Access Control Models and Technologies, Chantilly, VA, USA, 3–4 May 2001; Volume 172, pp. 3–9. [Google Scholar] [CrossRef]
- Eden, M.J.; Furley, P.A.; Mcgregor, D.F.M.; Milliken, W.; Ratter, J.A. Effect of forest clearance and burning on soil properties in northern Roraima, Braz. For. Ecol. Manag. 1991, 38, 283–290. [Google Scholar] [CrossRef]
- Tamura, S.; Oshima, T.; Yoshihara, K.; Kanazawa, A.; Yamada, T.; Inagaki, D.; Sato, T.; Yamamoto, N.; Shiozawa, M.; Morinaga, S. Clinical significance of STC1 gene expression in patients with colorectal cancer. Anticancer Res. 2011, 31, 325–329. [Google Scholar] [PubMed]
- Kempe, J. Biological control of bacterial wilt of potatoes: Attempts to induce resistance by treating tubers with bacteria. Plant Dis. 1983, 67, 499–503. [Google Scholar] [CrossRef]
- Champoiseau, P.G.; Momol, T.M. Bacterial wilt of tomato. Ralstonia solanacearum 2008, 12. [Google Scholar]
- Wang, Z.Y.; Thornton, C.R.; Kkershaw, M.J.; Debao, L.; Talbot, N.J. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 2003, 47, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Jenkinson, J.M.; Holcombe, L.J.; Soanes, D.M.; Veneault-Fourrey, C.; Bhambra, G.K.; Talbot, N.J. The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem. Soc. Trans. 2005, 33, 384–388. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples are available from the authors. |



| Test | Result | Test | Result |
|---|---|---|---|
| Cell morphology | Long rod | Gram stain | - |
| Oxidase | - | Contact enzyme | + |
| Citrate | - | V-P determination | - |
| Hydrogen sulfide | - | Lysine decarboxylase | - |
| Ornithine decarboxylase | - | Arginine dihydrolase | - |
| Nitrate | + | Starch hydrolysis | - |
| Urease | - | Lecithinase | - |
| Indole test | + | Cellobiose | - |
| Arabinose | - | Lactose | - |
| Sorbitol | + | Adonitol | - |
| Glucose | + | Maltose | + |
| Xylose | + | Trehalose | + |
| Mannitol | + | ONPG | - |
| Malonate | - | Methyl red | + |
| Grade | Disease Description |
|---|---|
| DI-0 | No visible symptoms |
| DI-1 | Up to 25% of leaves wilted |
| DI-2 | 25–50% of leaves wilted |
| DI-3 | 50–75% of leaves wilted |
| DI-4 | 75–100% of leaves wilted, the plants always died |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Fu, S.; Sun, X.; Li, P.; Wu, J.; Dong, T.; He, F.; Deng, Y. Identification of Cyclic Dipeptides from Escherichia coli as New Antimicrobial Agents against Ralstonia Solanacearum. Molecules 2018, 23, 214. https://doi.org/10.3390/molecules23010214
Song S, Fu S, Sun X, Li P, Wu J, Dong T, He F, Deng Y. Identification of Cyclic Dipeptides from Escherichia coli as New Antimicrobial Agents against Ralstonia Solanacearum. Molecules. 2018; 23(1):214. https://doi.org/10.3390/molecules23010214
Chicago/Turabian StyleSong, Shihao, Shuna Fu, Xiuyun Sun, Peng Li, Ji’en Wu, Tingyan Dong, Fei He, and Yinyue Deng. 2018. "Identification of Cyclic Dipeptides from Escherichia coli as New Antimicrobial Agents against Ralstonia Solanacearum" Molecules 23, no. 1: 214. https://doi.org/10.3390/molecules23010214




