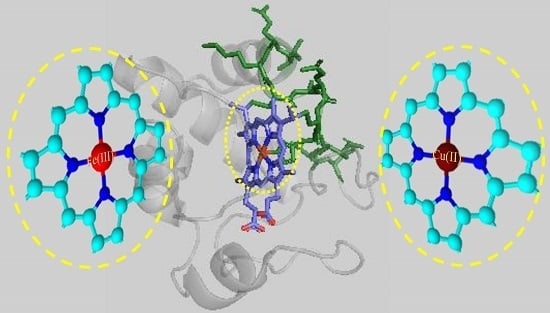
Structure and Catalysis of Fe(III) and Cu(II) Microperoxidase-11 Interacting with the Positively Charged Interfaces of Lipids
Abstract
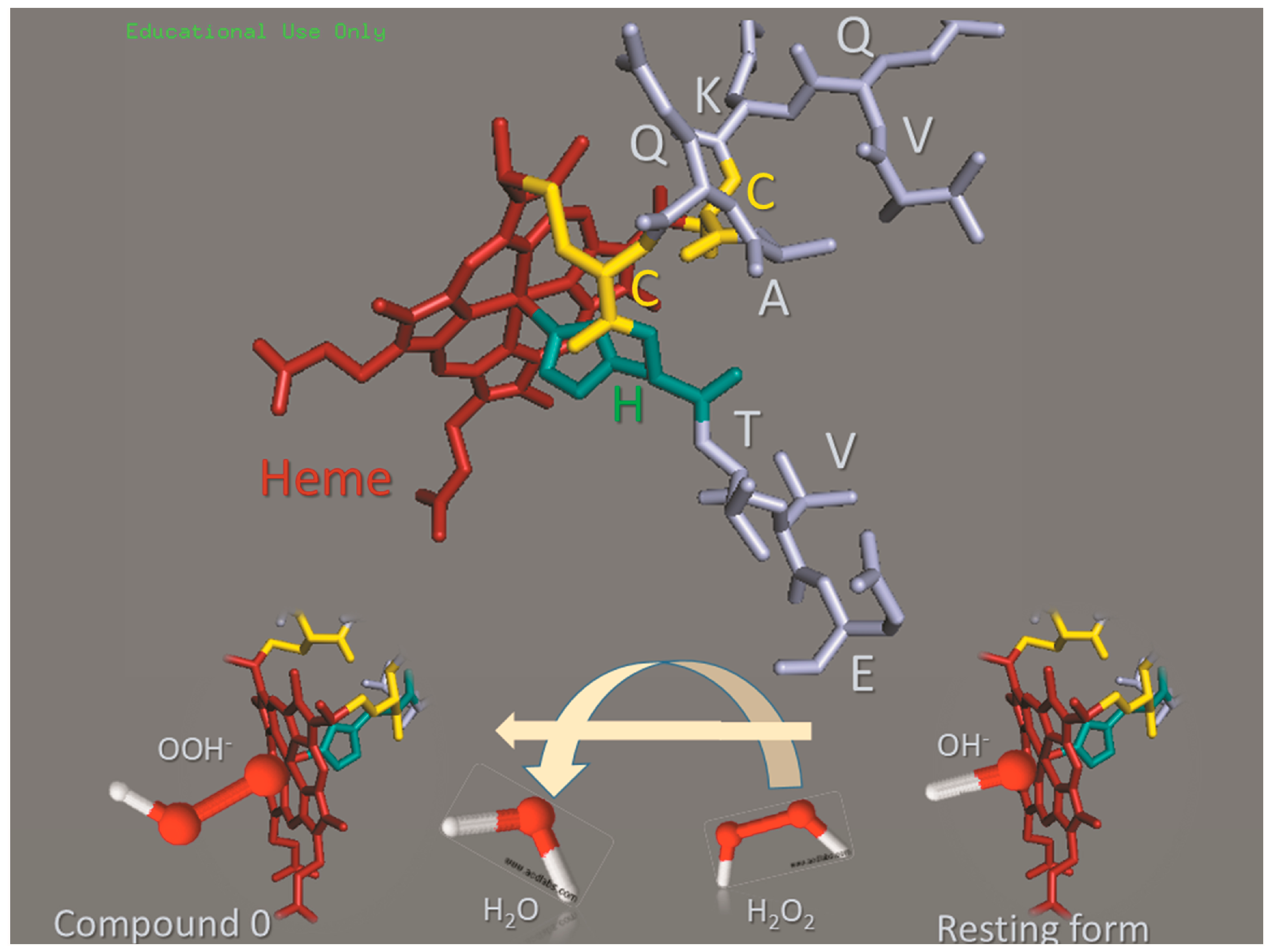
:1. Introduction
2. Results
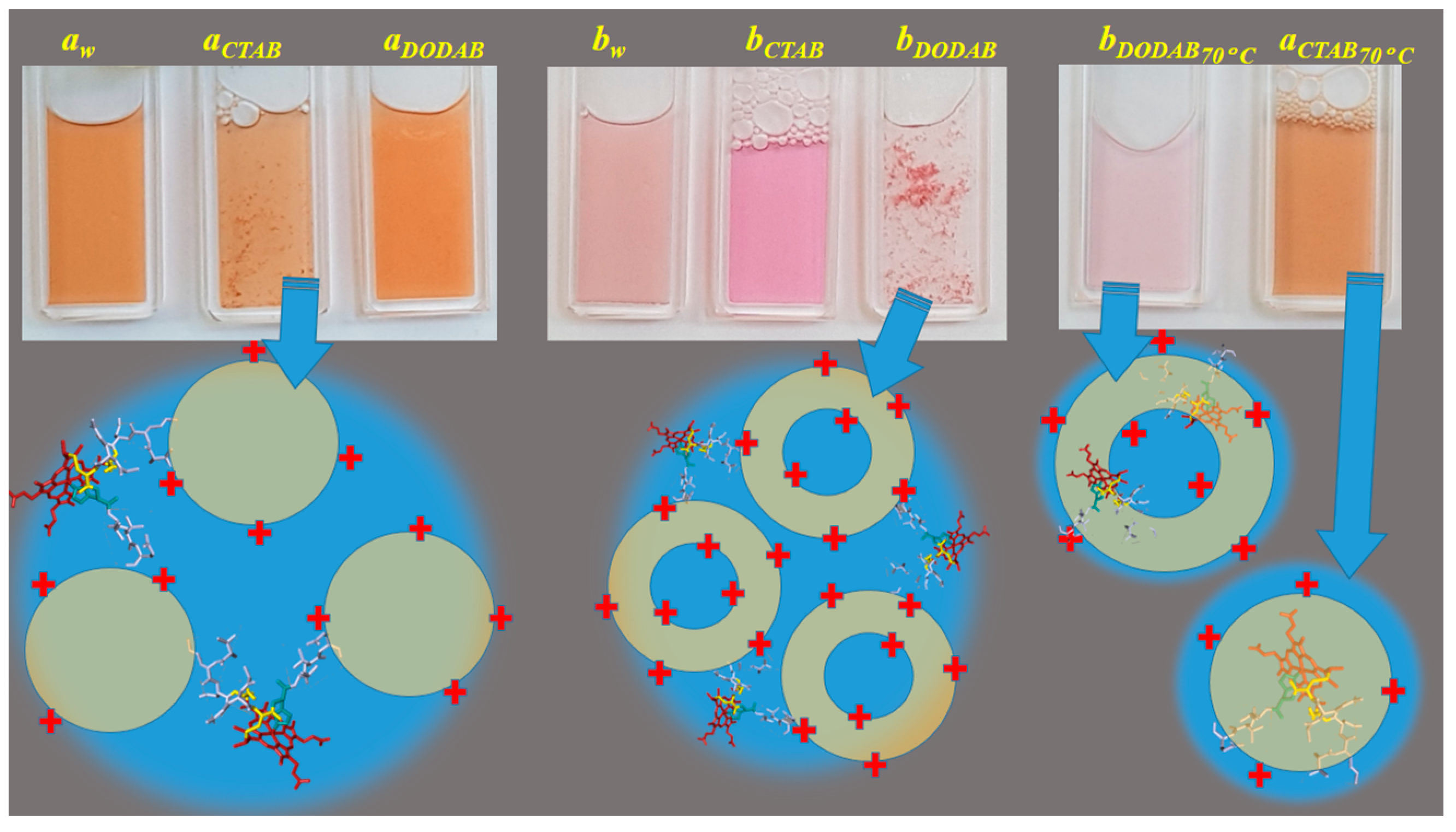
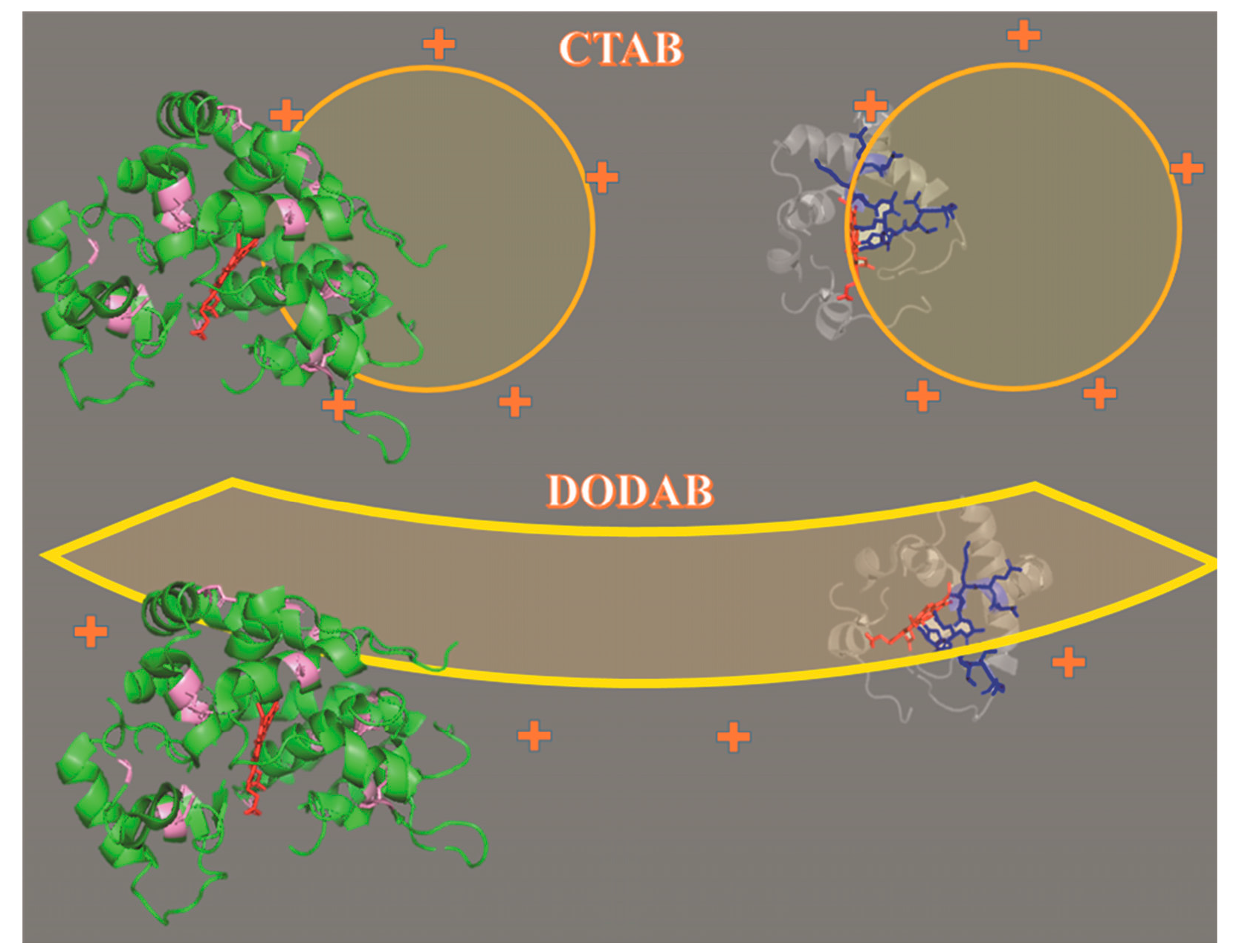
2.1. Cu(II) and Fe(III)MP-11 in Homogeneous and Heterogeneous Media
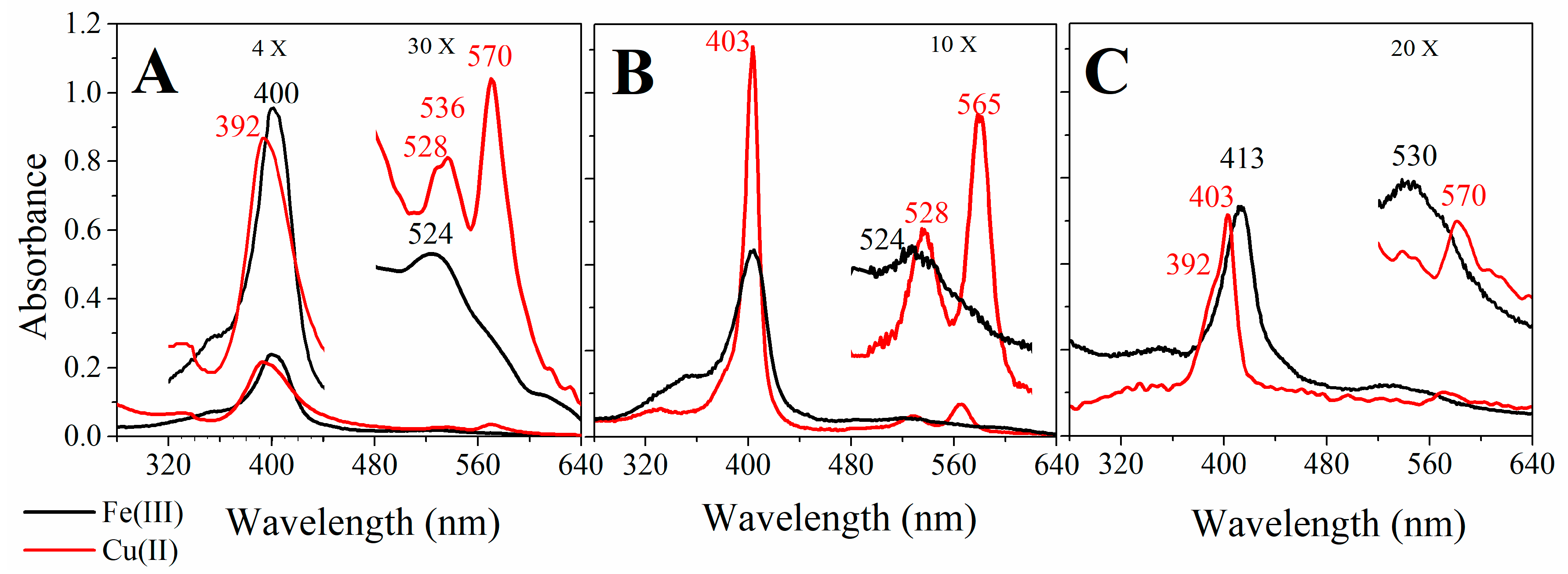
2.2. UV-Visible Spectral Features of Cu(II) and Fe(III)MP-11 in Homogeneous and Heterogeneous Media
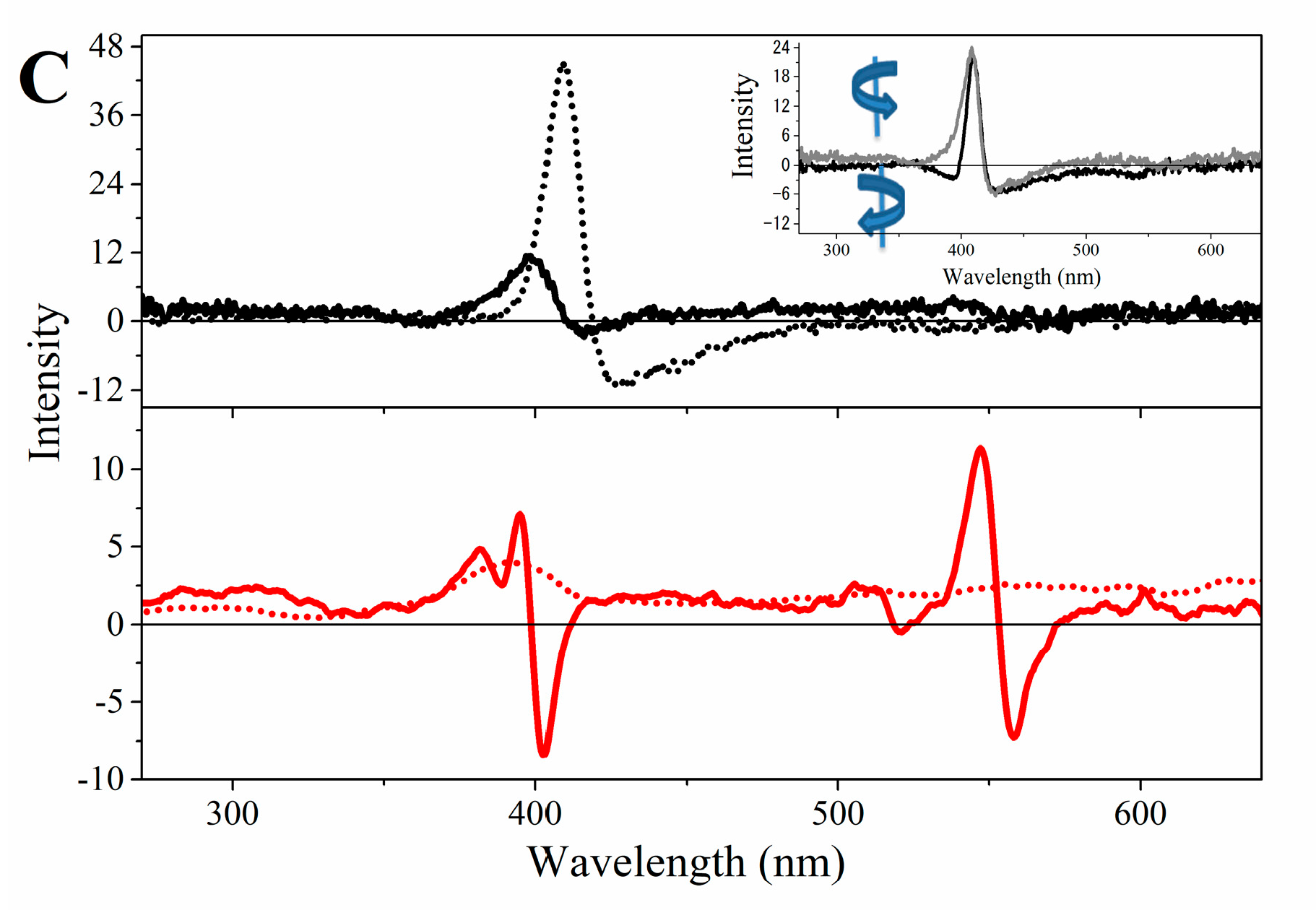
2.3. Magnetic Circular Dichroism Spectral Features of Cu(II) and Fe(III) MP-11 in Homogeneous and Heterogeneous Media
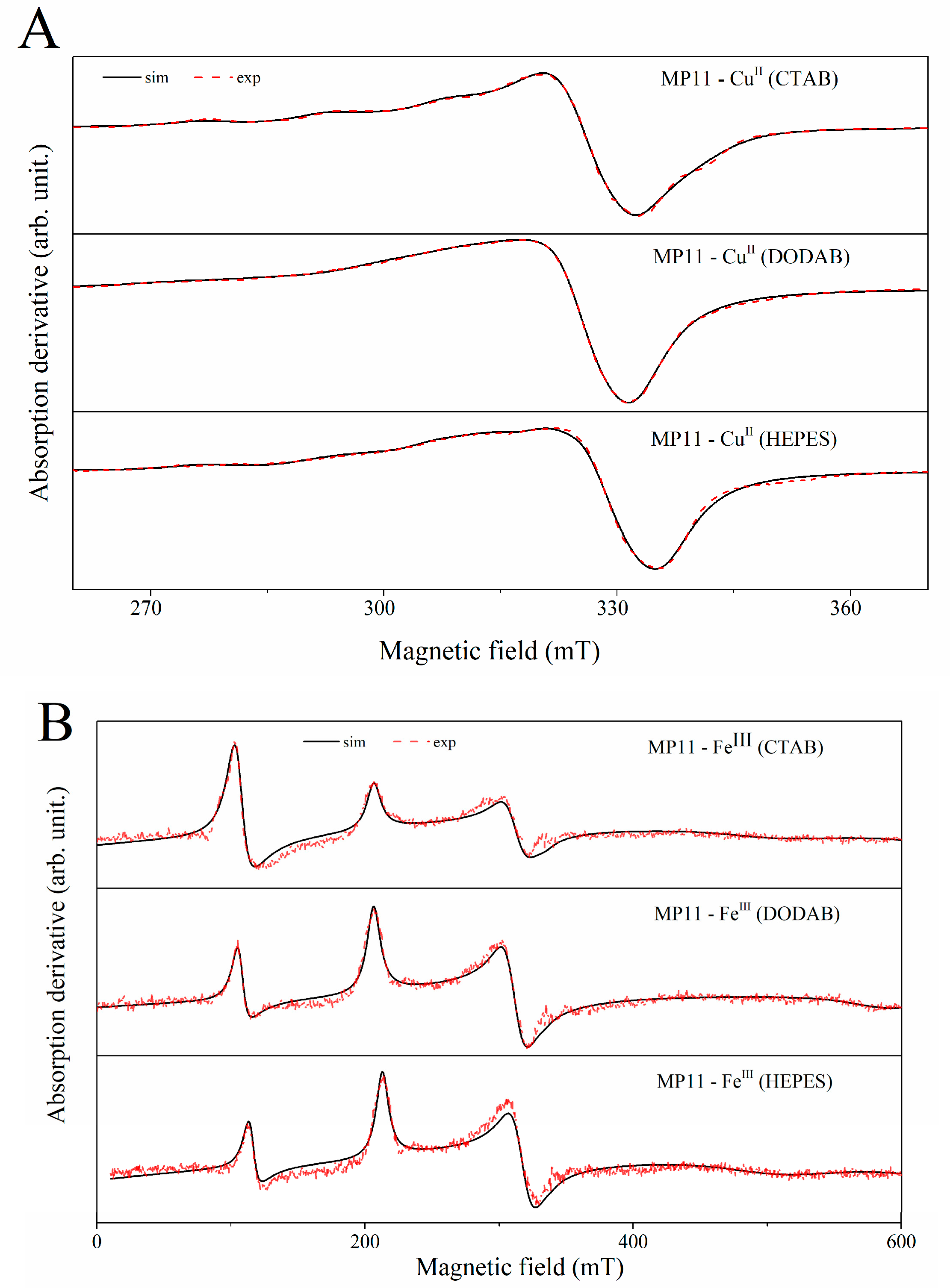
2.4. EPR Spectral Characteristics of Cu(II) and Fe(III) MP-11 in Homogeneous and Heterogeneous Media
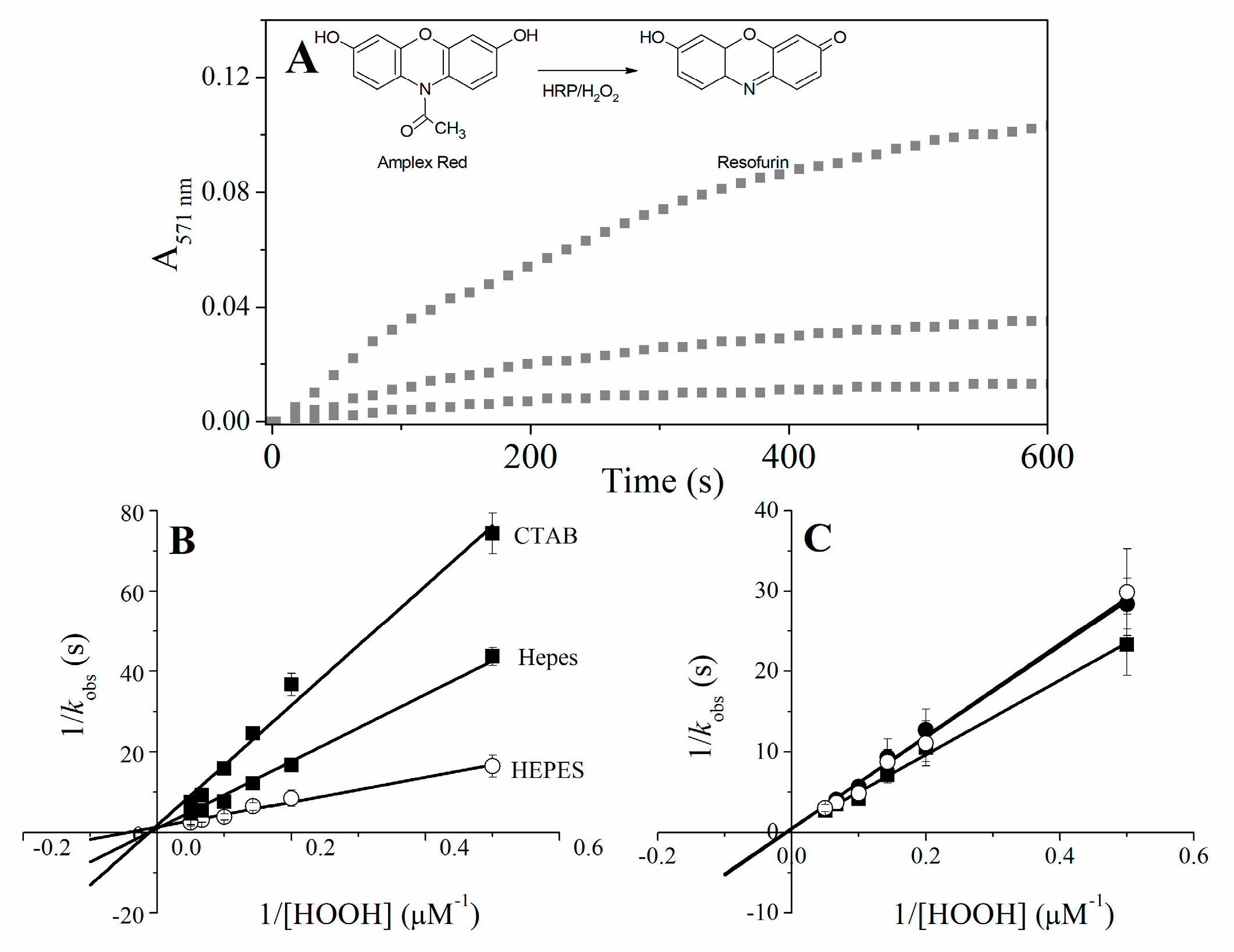
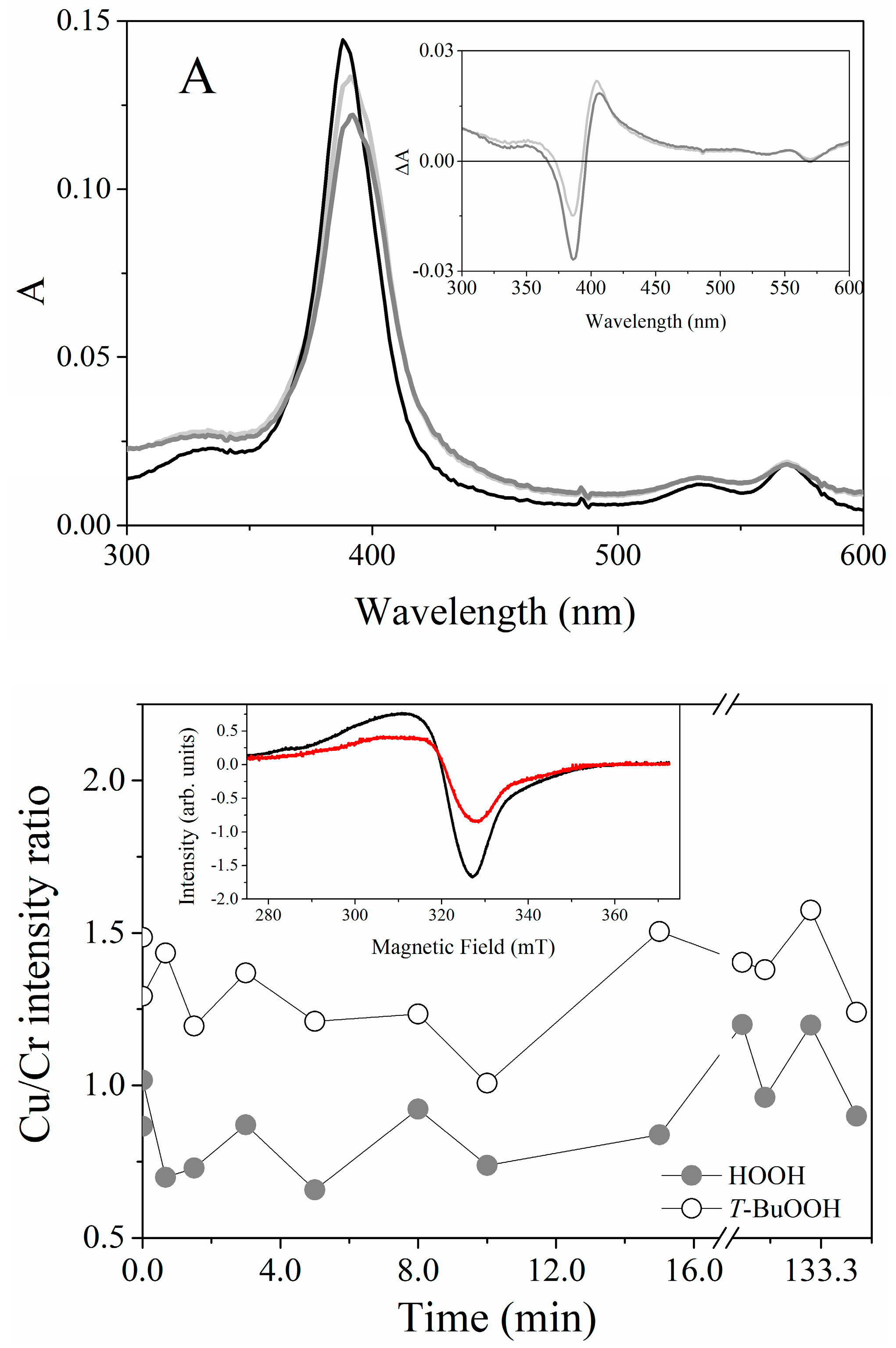
2.5. Catalytic Properties of Cu(II) and Fe(III)MP-11 Homogeneous and Heterogeneous Media Using Amplex® Red
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Preparation of Cu(II)MP-11 [42]
4.3. Preparation of the CTAB Micelles and Large DODAB Vesicles
4.4. Zeta-Potential Measurements
4.5. Determination of UV-Visible Optical Spectra
4.6. Magnetic Circular Dichroism (MCD)
4.7. Determination of Heme Fe(III) and Cu(II) EPR Spectra
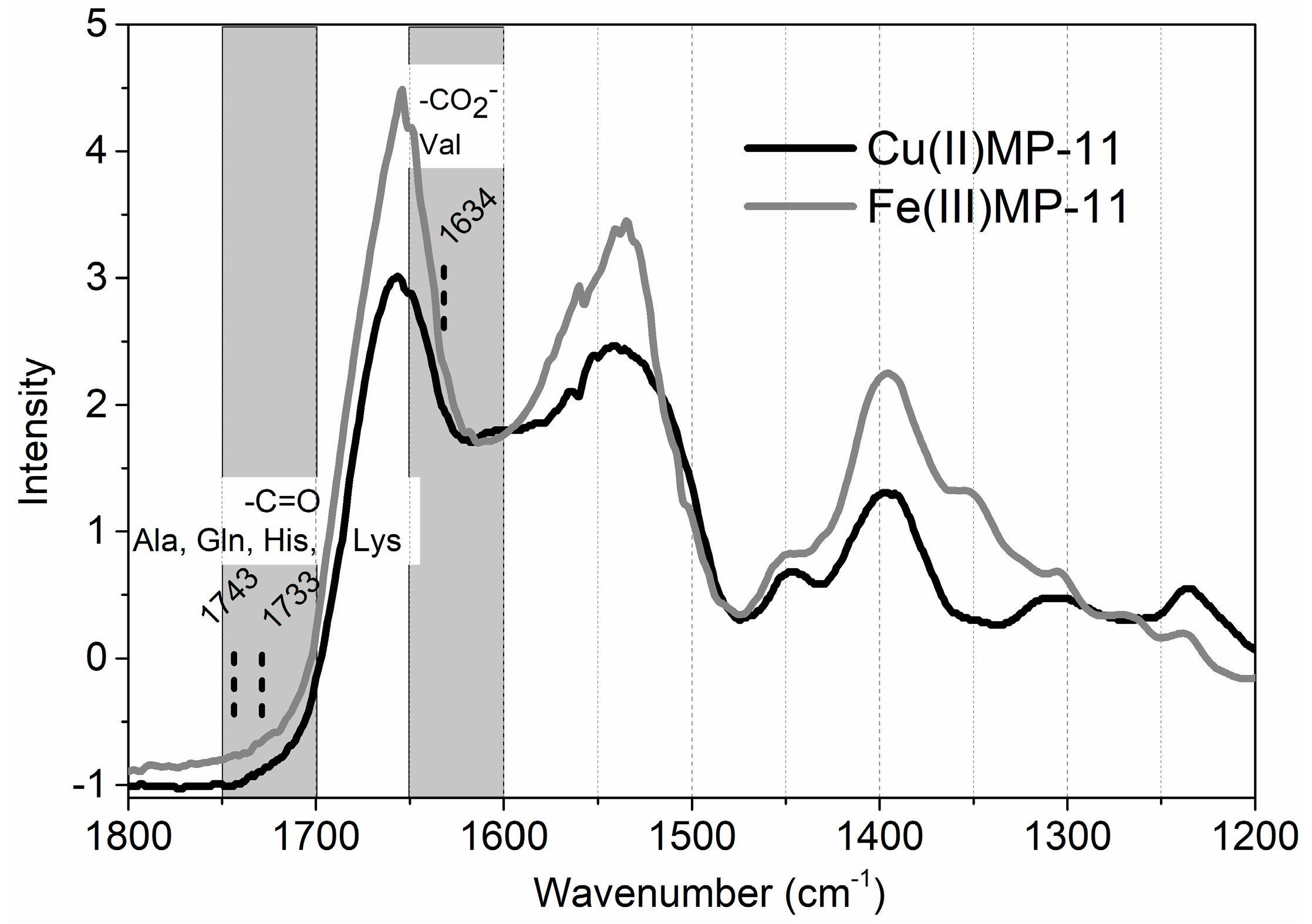
4.8. FTIR Spectroscopy
4.9. Amplex® Red Oxidation
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Aron, J.; Baldwin, D.A.; Marques, H.M.; Pratt, J.M.; Adams, P.A. Hemes and hemoproteins. J. Inorg. Biochem. 1986, 27, 227–243. [Google Scholar] [CrossRef]
- Wang, T.; Liu, J.; Ren, J.; Wang, J.; Wang, E. Mimetic biomembrane—AuNPs—Graphene hybrid as matrix for enzyme immobilization and bioelectrocatalysis study. Talanta 2015, 143, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Kleingardner, E.C.; Asher, W.B.; Bren, K.L. Efficient and Flexible Preparation of Biosynthetic Microperoxidases. Biochemistry 2017, 56, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Thöny-Meyer, L. Biosynthesis of artificial microperoxidases by exploiting the secretion and cytochrome c maturation apparatuses of Escherichia coli. Proc. Natl. Acad. Soc. USA 2004, 101, 12830–12835. [Google Scholar] [CrossRef] [PubMed]
- Munro, O.Q.; Marques, H.M. Heme−Peptide Models for Hemoproteins. 1. Solution Chemistry of N-Acetylmicroperoxidase-8. Inorg. Chem. 1996, 35, 3752–3767. [Google Scholar] [CrossRef] [PubMed]
- Carraway, A.D.; Povlock, S.L.; Houston, M.L.; Johnston, D.S.; Peterson, J. Monomeric ferric heme peptide derivatives: Model systems for hemoproteins. J. Inorg. Biochem. 1995, 60, 267–276. [Google Scholar] [CrossRef]
- Veeger, C. Does P450-type catalysis proceed through a peroxo-iron intermediate? A review of studies with microperoxidase. J. Inorg. Biochem. 2002, 91, 35–45. [Google Scholar] [CrossRef]
- Peterson, J.; Saleem, M.M.M.; Silver, J.; Wilson, M.T.; Morrison, I.E.G. On the Preparation and Mössbauer Properties of Sonic Heme Peptides of Cytochrome c. J. Inorg. Biochem. 1983, 19, 165–178. [Google Scholar] [CrossRef]
- Dunford, H. On the function and mechanism of action of peroxidases. Coord. Chem. Rev. 1976, 19, 187–251. [Google Scholar] [CrossRef]
- Dunford, H.B. Horseradish peroxidase: Structure and kinetic properties. In Peroxidases in Chemistry and Biology; Everse, J., Everse, K.E., Grisham, M.B., Eds.; CRC Press: Boca Raton, FL, USA, 1991; Volume 2, pp. 1–24. [Google Scholar]
- Nantes, I.L.; Durán, N.; Pinto, S.M.S.; da Silva, F.B.; de Souza, J.S.; Isoda, N.; Luz, R.A.S.; de Oliveira, T.G.; Fernandes, V.G. Modulation of the catalytic activity of porphyrins by lipid-and surfactant-containing nanostructures. J. Braz. Chem. Soc. 2011, 22, 1621–1633. [Google Scholar] [CrossRef]
- Prieto, T.; Marcon, R.O.; Prado, F.M.; Caires, A.C.F.; Di Mascio, P.; Brochsztain, S.; Nascimento, O.R.; Nantes, I.L. Reaction route control by microperoxidase-9/CTAB micelle ratios. Phys. Chem. Chem. Phys. 2006, 8, 1963. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yuri, I.; Gan, X.; Suzuki, I.; Li, G. Electrochemical study of the effect of nano-zinc oxide on microperoxidase and its application to more sensitive hydrogen peroxide biosensor preparation. Biosens. Bioelectron. 2007, 22, 1600–1604. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.C.; Prieto, T.; Prado, F.M.; Trindade, F.J.; Nunes, G.L.C.; dos Santos, J.G.; Di Mascio, P.; Castro, F.L.; Fernandes, G.J.; Fernandes, V.J.; et al. Peroxidase Catalytic Cycle of MCM-41-Entrapped Microperoxidase-11 as a Mechanism for Phenol Oxidation. J. Nanosci. Nanotechnol. 2007, 7, 3643–3652. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; Kielb, P.; Rustam, L.; Fischer, A.; Weidinger, I.M.; Wollenberger, U. Bioelectrocatalytic Reduction of Hydrogen Peroxide A. Bioelectrocataliyic Reduction of Hydrogen Peroxide by Microperoxidase-11 Immobilized on Mesoporous Antimony-Doped Tin Oxide. ChemElectroyChem 2017, 4, 913. [Google Scholar]
- Gonga, C.; Shena, Y.; Chena, J.; Songa, Y.; Chena, S.; Songa, Y.; Wanga, L. Microperoxidase-11@PCN-333 (Al)/three-dimensional macroporous carbon electrode for sensing hydrogen peroxide. Sens. Actuators B Chem. 2017, 239, 890–897. [Google Scholar] [CrossRef]
- Riposati, A.; Prieto, T.; Shida, C.S.; Nantes, I.L.; Nascimento, O.R. Low spin states of microperoxidases produced by inter- and intra-peptide chain sixth ligands: Effect of pH and the oligopeptide type. J. Inorg. Biochem. 2006, 100, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Lisi, G.P.; Collins, D.P.; Dawson, J.H.; Pletneva, E.V. Redox-dependent stability, protonation, and reactivity of cysteine-bound heme proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E306–E315. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Tsai, A.L.; Heldt, J.; Palmer, G.; Van Wart, H.E. Temperature- and pH-dependent changes in the coordination sphere of the heme c group in the model peroxidase N-alpha-acetyl microperoxidase-8. J. Biol. Chem. 1992, 267, 15310–15318. [Google Scholar] [PubMed]
- Bieza, S.A.; Boubeta, F.; Feis, A.; Smulevich, G.; Estrin, D.A.; Boechi, L.; Bari, S.E. Reactivity of Inorganic Sulfide Species toward a Heme Protein Model. Inorg. Chem. 2015, 54, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Silveira, C.M.; Castro, M.A.; Dantas, J.M.; Salgueiro, C.; Murgida, D.H.; Todorovic, S. Structure, electrocatalysis and dynamics of immobilized cytochrome PccH and its microperoxidase. Phys. Chem. Chem. Phys. 2017, 19, 8908–8918. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, Q.; Su, B. Adsorption of Microperoxidase-11 in Vertical Silica Mesochannels and Electrochemical Investigation of Its Electron Transfer Properties. Electrochim. Acta 2015, 161, 290–296. [Google Scholar] [CrossRef]
- Ascenzi, P.; Leboffe, L.; Santucci, R.; Coletta, M. Ferric microperoxidase-11 catalyzes peroxynitrite isomerization. J. Inorg. Biochem. 2015, 144, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Rusvai, E.; Végh, M.; Kramer, M.; Horváth, I. Hydroxylation of aniline mediated by heme-bound oxy-radicals in a heme peptide model system. Biochem. Pharmacol. 1988, 37, 4574–4577. [Google Scholar] [CrossRef]
- Graça, J.S.; de Oliveira, R.F.; de Moraes, M.L.; Ferreira, M. Amperometric glucose biosensor based on layer-by-layer films of microperoxidase-11 and liposome-encapsulated glucose oxidase. Bioelectrochemistry 2014, 96, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Han, L.; Wang, T.; Hong, W.; Liu, Y.; Wang, E. Enzyme Immobilization and Direct Electrochemistry Based on a New Matrix of Phospholipid-Monolayer-Functionalized Graphene. Chem. Asian J. 2012, 7, 2824–2829. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zhou, Q.; Gu, Z.; Gu, X.; Zhao, L.; Li, Y.; Zheng, J. Hydrogen peroxide biosensor based on microperoxidase-11 immobilized in a silica cavity array electrode. Talanta 2013, 107, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhou, J.; Li, S.; Zhang, X.; Huang, D.; He, Y.; Wang, M.; Yang, G.; Shen, Y. Hydrogen peroxide biosensor based on microperoxidase-11 immobilized on flexible MWCNTs-BC nanocomposite film. Talanta 2015, 131, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Kleingardner, J.G.; Kandemir, B.; Bren, K.L. Hydrogen Evolution from Neutral Water under Aerobic Conditions Catalyzed by Cobalt Microperoxidase-11. J. Am. Chem. Soc. 2014, 136, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Marques, H.M.; Wilkins, R.G.; Klapper, M.H.; Giacometti, G.; Merli, A.; Brunori, M.; Canters, G.W.; Van Dongen, W.M.A.M.; Rietjens, I.M.C.M. Insights into porphyrin chemistry provided by the microperoxidases, the haempeptides derived from cytochrome c. Dalton Trans. 2007, 29, 4371. [Google Scholar] [CrossRef] [PubMed]
- Verbaro, D.; Hagarman, A.; Kohli, A.; Schweitzer-Stenner, R. Microperoxidase 11: A model system for porphyrin networks and heme–protein interactions. JBIC J. Biol. Inorg. Chem. 2009, 14, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Zoppellaro, G.; Bren, K.L.; Ensign, A.A.; Harbitz, E.; Kaur, R.; Hersleth, H.-P.; Ryde, U.; Hederstedt, L.; Andersson, K.K. Review: Studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-Histidine and histidine-methionine axial iron coordination. Biopolymers 2009, 91, 1064–1082. [Google Scholar] [CrossRef]
- Kadnikova, E.N.; Kostić, N.M. Effects of the Environment on Microperoxidase-11 and on Its Catalytic Activity in Oxidation of Organic Sulfides to Sulfoxides. J. Org. Chem. 2003, 68, 2600–2608. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Chi, Y.S.J. Vibrational and Electronic Spectroscopic Characterizations of Amino Acid-Metal Complexes. Korean Soc. Appl. Biol. Chem. 2010, 53, 821–825. [Google Scholar] [CrossRef]
- Araujo-Chaves, J.C.; Santos, C.G.; Miranda, E.G.A.; Arantes, J.T.; Nantes, I.L. Free-Base and Metal Complexes of 5,10,15,20-Tetrakis(NMethyl Pyridinium L)Porphyrin: Catalytic and Therapeutic Properties, Phthalocyanines and Some Current Applications; Yilmaz, Y., Ed.; InTech: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef]
- Gorris, H.H.; Walt, D.R. Mechanistic Aspects of Horseradish Peroxidase Elucidated through Single-Mo lecule Studies. J. Am. Chem. Soc. 2009, 131, 6277–6282. [Google Scholar] [CrossRef] [PubMed]
- Prieto, T.; Nascimento, O.R.; Tersariol, I.L.S.; Faljoni-Alario, A.; Nantes, I.L. Microperoxidase-8 Associated to CTAB Micelles: A New Catalyst with Peroxidase Activity. J. Phys. Chem. B 2004, 108, 11124–11132. [Google Scholar] [CrossRef]
- Bonini, M.G.; Gabel, S.A.; Ranguelova, K.; Stadler, K.; Derose, E.F.; London, R.E.; Mason, R.P. Direct Magnetic Resonance Evidence for Peroxymonocarbonate Involvement in the Cu,Zn-Superoxide Dismutase Peroxidase Catalytic Cycle. J. Biol. Chem. 2009, 284, 14618–14627. [Google Scholar] [CrossRef] [PubMed]
- Nims, R.W.; Prough, R.A.; Lubet, R.A. Cytosol-Mediated Reduction of Resorufin: A Method for Measuring Quinone Oxidoreductase. Arch. Biochem. Biophys. 1984, 229, 459–465. [Google Scholar] [CrossRef]
- Song, P.; Ruan, M.; Sun, X.; Zhang, Y.; Xu, W. Theoretical Study of Resorufin Reduction Mechanism by NaBH4. J. Phys. Chem. B 2014, 118, 10224–10231. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Chaves, J.C.; Yokomizo, C.H.; Kawai, C.; Mugnol, K.C.U.; Prieto, T.; Nascimento, O.R.; Nantes, I.L. Towards the mechanisms involved in the antioxidant action of MnIII [meso-tetrakis(4-N-methyl pyridinium) porphyrin] in mitochondria. J. Bioenerg. Biomembr. 2011, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findlay, M.C.; Dickinson, L.C.; Chien, J.C.W. Copper-cytochrome c. J. Am. Chem. Soc. 1977, 99, 5168–5173. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, L.C.; Chien, J.C.W. Cobalt-cytochrome c. I. Preparation, properties, and enzymic activity. Biochemistry 1975, 14, 3526–3534. [Google Scholar] [CrossRef] [PubMed]
- Dearden, L.V.; Woolley, E.M. Osmotic coefficients of alkyltrimethylammonium bromides in water and in aqueous sodium bromide solutions at 55. degree. C. J. Phys. Chem. 1987, 91, 2404–2408. [Google Scholar] [CrossRef]
- Sobral, C.N.C.; Soto, M.A.; Carmona-Ribeiro, A.M. Characterization of DODAB/DPPC vesicles. Chem. Phys. Lipids 2008, 152, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, D.B.; Rapuano, R.; Lessa, M.M.; Carmona-Ribeiro, A.M. Counterion Effects on Properties of Cationic Vesicles. Langmuir 1998, 14, 7387–7391. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Carmona, A.M. Biomimetic nanoparticles: Preparation, characterization and biomedical applications. Int. J. Nanomed. 2010, 5, 249–259. [Google Scholar] [CrossRef]
- Ohlsson, P.I.; Paul, K.G. The molar absorptivity of horseradish peroxidase. Acta Chem. Scand. B 1976, 30, 373–375. [Google Scholar] [CrossRef]
- Zielonka, J.; Zielonka, M.; Sikora, A.; Adamus, J.; Joseph, J.; Hardy, M.; Ouari, O.; Dranka, B.P.; Kalyanaraman, B. Global Profiling of Reactive Oxygen and Nitrogen Species in Biological Systems: High-throughput real-time analyses. J. Biol. Chem. 2012, 287, 2984–2995. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |


| MP-11 | ξ (mV) |
|---|---|
| Cu(II) MP-11 | −27.8 ± 3 |
| Fe(III) MP-11 | −13.1 ± 5 |
| CTAB micelles | +50.5 ± 2 |
| Cu(II)MP-11/CTAB micelles | +34.3 ± 1 |
| Fe(III)MP-11/CTAB micelles | +46.1 ± 3 |
| DODAB vesicles | +65.7 ± 6 |
| Cu(II)MP-11/DODAB vesicles | +61.6 ± 7 |
| Fe(III)MP-11/DODAB vesicles | +57.1 ± 4 |
| Cu(II)MP-11 | Fe(III)MP-11 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Medium | gx | gy | gz | Ax (MHz) | Ay (MHz) | Az (MHz) | ΔH (mT) | % | gx | gy | gz | ΔH (mT) | % | |
| Sp.1 | HEPES | 2.04 | 2.11 | 2.25 | 60 | 108 | 482 | 8.5 | 44 | 5.89 | 2 | 12.5 | 0.09 | |
| CTAB | 2.05 | 2.08 | 2.24 | 38 | 56 | 466 | 2 | 63 | 5.95 | 2 | 18.2 | 0.54 | ||
| DODAB | 2.04 | 2.09 | 2.31 | 54 | 70 | 356 | 17 | 61 | 5.88 | 2 | 12.7 | 0.09 | ||
| Sp.2 | HEPES | 2.04 | 2.06 | 2.19 | 11 | 53 | 237 | 11 | 56 | 1.32 | 2.13 | 3.18 | 10.8 | 99.91 |
| CTAB | 2.02 | 2.02 | 2.11 | 33 | 33 | 367 | 9 | 37 | 1.32 | 2.13 | 3.17 | 12.2 | 99.46 | |
| DODAB | 2.04 | 2.07 | 2.14 | 60 | 18 | 260 | 8 | 39 | 1.14 | 2.14 | 3.18 | 11.9 | 99.91 | |
| HRP | Fe(III)MP-11 | |||||
|---|---|---|---|---|---|---|
| Kcat (s−1) | Km (μM) | Kcat/Km (μM−1·s−1) | Kcat (s−1) | Km (μM) | Kcat/Km (μM−1·s−1) | |
| HEPES | 10.53 | 109.90 ± 11.0 | 0.096 | 0.75 | 23.80 ± 2.2 | 0.031 |
| CTAB | 7.84 | 128.20 ± 5.4 | 0.061 | 1.00 | 71.43 ± 1.9 | 0.014 |
| DODAB | 8.88 | 151.51 ± 10.3 | 0.058 | 0.56 | 83.33 ± 5.71 | 0.0067 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prieto, T.; Santana, V.; Britto, A.M.M.; Araujo-Chaves, J.C.; R. Nascimento, O.; L. Nantes-Cardoso, I. Structure and Catalysis of Fe(III) and Cu(II) Microperoxidase-11 Interacting with the Positively Charged Interfaces of Lipids. Molecules 2017, 22, 1212. https://doi.org/10.3390/molecules22081212
Prieto T, Santana V, Britto AMM, Araujo-Chaves JC, R. Nascimento O, L. Nantes-Cardoso I. Structure and Catalysis of Fe(III) and Cu(II) Microperoxidase-11 Interacting with the Positively Charged Interfaces of Lipids. Molecules. 2017; 22(8):1212. https://doi.org/10.3390/molecules22081212
Chicago/Turabian StylePrieto, Tatiana, Vinicius Santana, Adrianne M. M. Britto, Juliana C. Araujo-Chaves, Otaciro R. Nascimento, and Iseli L. Nantes-Cardoso. 2017. "Structure and Catalysis of Fe(III) and Cu(II) Microperoxidase-11 Interacting with the Positively Charged Interfaces of Lipids" Molecules 22, no. 8: 1212. https://doi.org/10.3390/molecules22081212