Co-Transcriptional Folding and Regulation Mechanisms of Riboswitches
Abstract
:1. Introduction
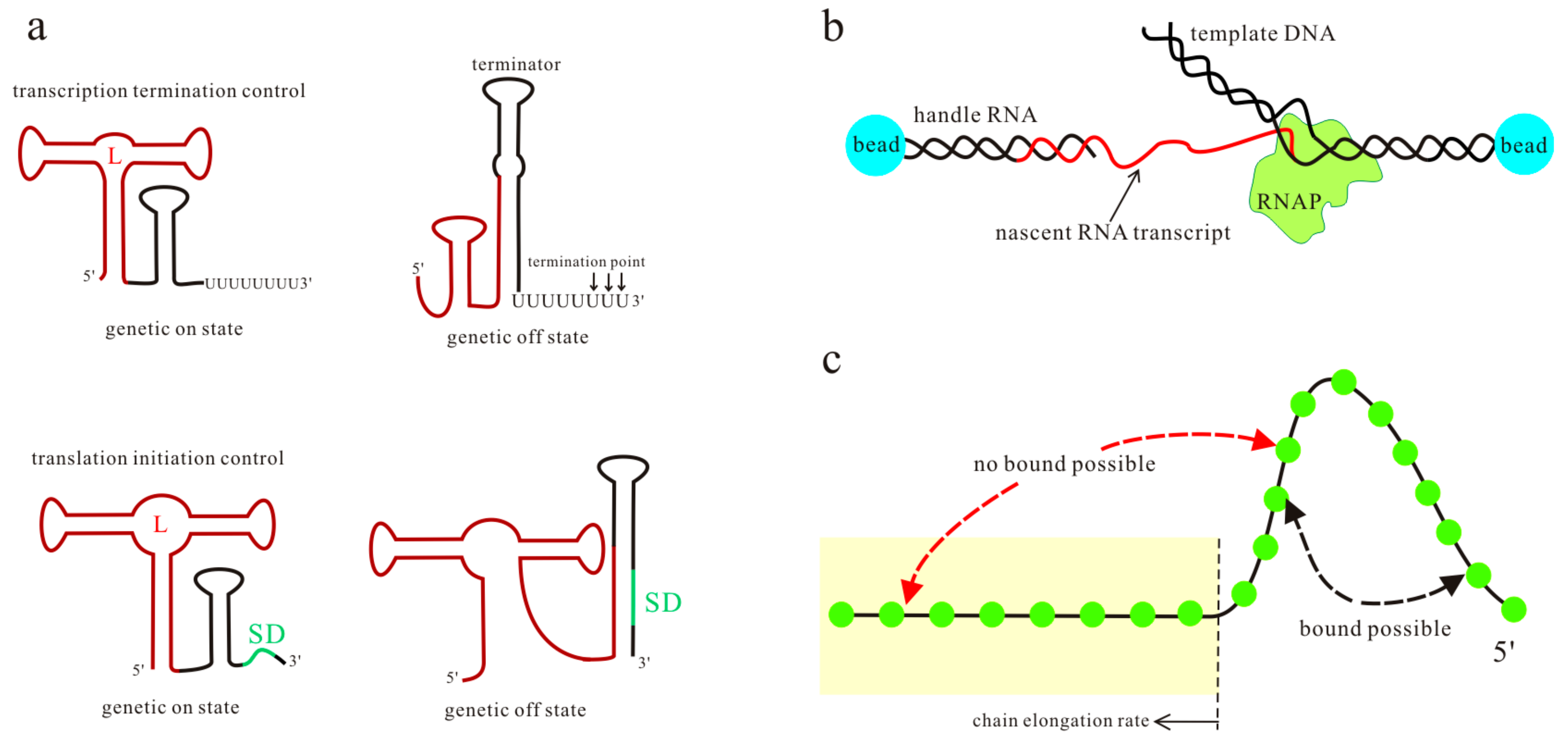
2. The Systematic Co-Transcriptional Folding Theory
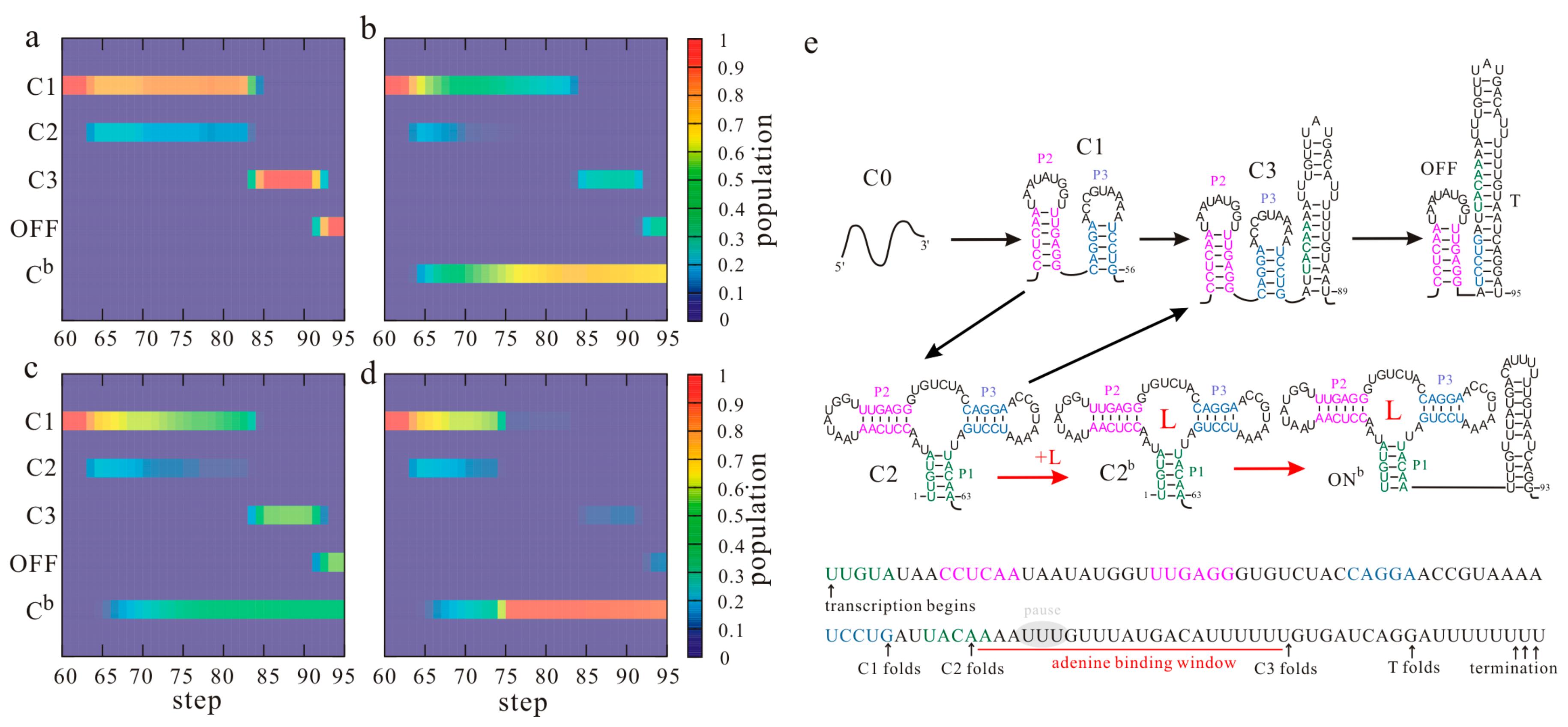
3. The Kinetic Regulation Regime of the pbuE and metF Riboswitches
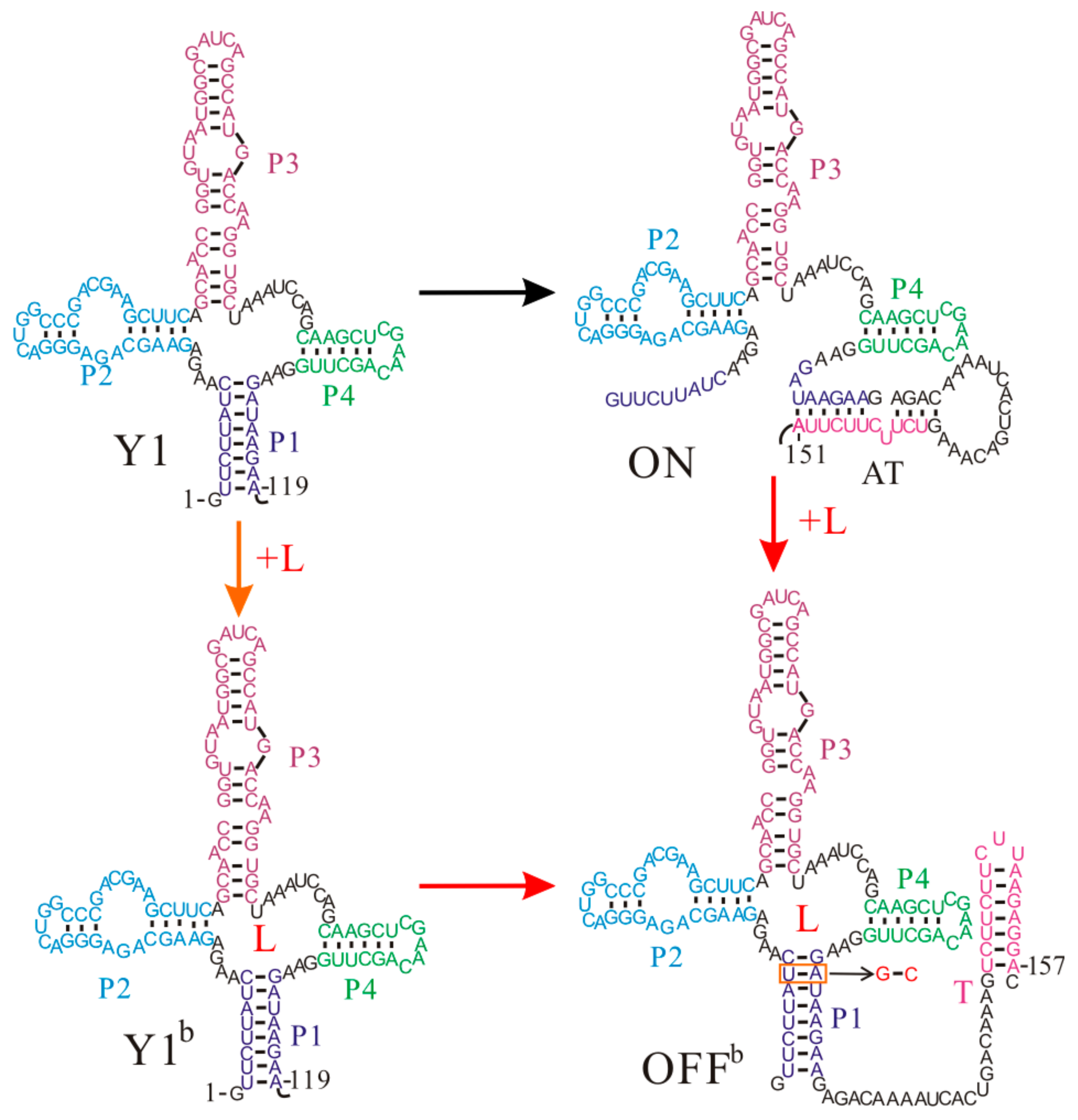
4. The Association of Thermodynamic and Kinetic Regulation Regimes of the yitJ Riboswitch
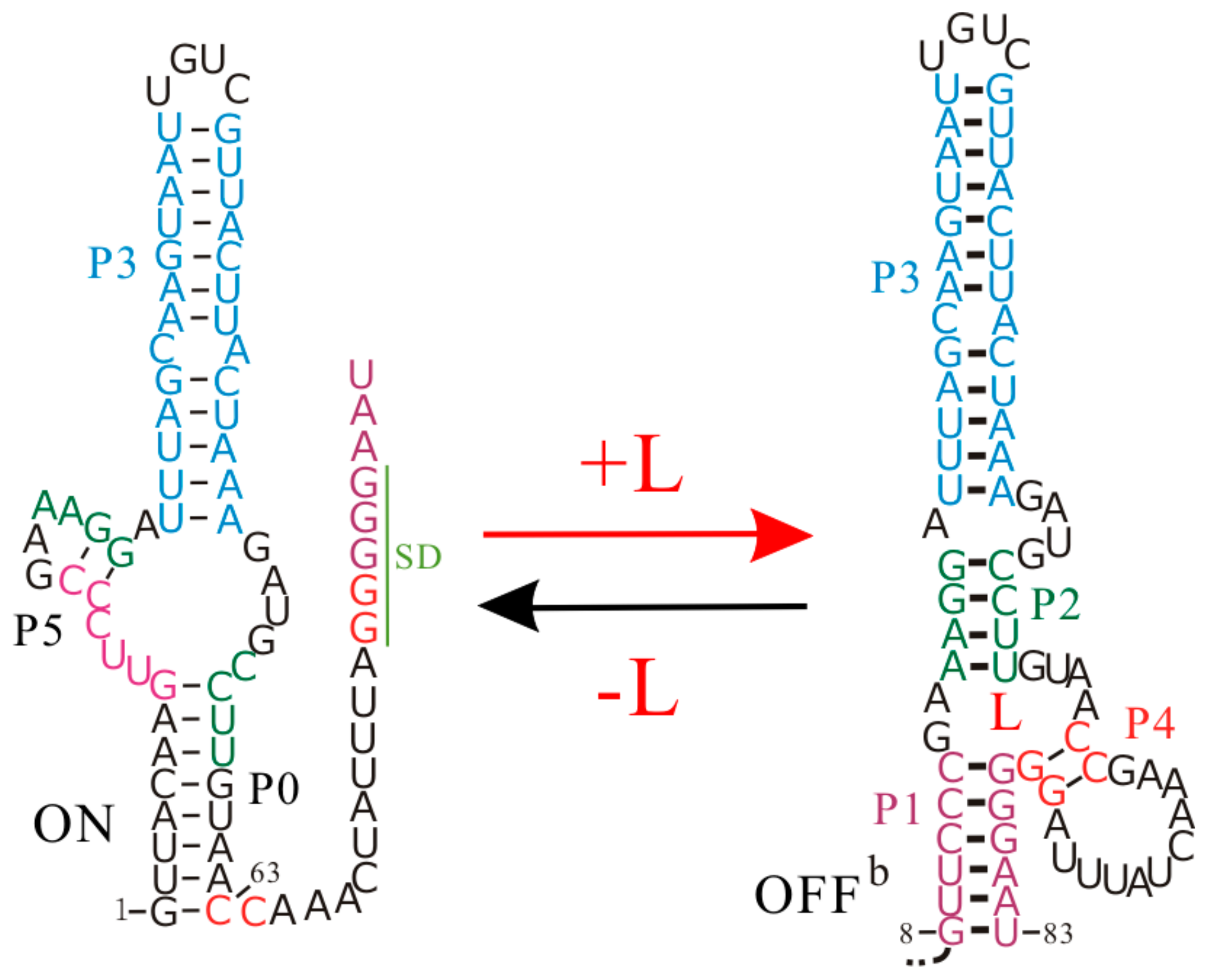
5. The Thermodynamic Regulation Regime of the SMK Riboswitch
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Reining, A.; Nozinovic, S.; Schlepckow, K.; Buhr, F.; Fürtig, B.; Schwalbe, H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 2013, 499, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-C.C.; Thirumalai, D. Kinetics of allosteric transitions in S-adenosylmethionine riboswitch are accurately predicted from the folding landscape. J. Am. Chem. Soc. 2013, 135, 16641–16650. [Google Scholar] [CrossRef] [PubMed]
- Perdrizet, G.A., II; Artsimovitch, I.; Furman, R.; Sosnick, T.R.; Pan, T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl. Acad. Sci. USA 2012, 109, 3323–3328. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Boese, B.; Barrick, J.E.; Winkler, W.C.; Breaker, R.R. Riboswitches Control Fundamental Biochemical Pathways in Bacillus subtilis and Other Bacteria. Cell 2003, 113, 577–586. [Google Scholar] [CrossRef]
- Winkler, W.C.; Nahvi, A.; Sudarsan, N.; Barrick, J.E.; Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003, 10, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Ramesh, A.; Zhou, Z.; Weinberg, Z.; Vallery, T.; Winkler, W.C.; Breaker, R.R. Bacterial Riboswitches Cooperatively Bind Ni2+ or Co2+ Ions and Control Expression of Heavy Metal Transporters. Mol. Cell 2015, 57, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Huang, L.; Patel, D.J. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 2009, 458, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Mellin, J.R.; Koutero, M.; Dar, D.; Nahori, M.-A.; Sorek, R.; Cossart, P. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 2014, 345, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Abbas, C.A.; Sibirny, A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005, 59, 487–517. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.M.; Hamelberg, D. Atomistic basis for the on-off signaling mechanism in SAM-II riboswitch. Nucleic Acids Res. 2010, 38, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ding, F.; Chowdhury, A.; Pradhan, V.; Tomsic, J.; Holmes, W.M.; Henkin, T.M.; Ke, A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 2010, 404, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004, 11, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Breaker, R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem 2003, 4, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Riboswitches: The oldest mechanism for the regulation of gene expression? TRENDS Genet. 2004, 20, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Wang, Y.; Zhang, W. Kinetic regulation mechanism of pbuE riboswitch. J. Chem. Phys. 2015, 142, 15103. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Breaker, R.R. Purine sensing by riboswitches. Biol. Cell 2008, 100, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wickiser, J.K.; Winkler, W.C.; Breaker, R.R.; Crothers, D.M. The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Mol. Cell 2005, 18, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Zhao, Y.; Chen, C.; Xiao, Y. Role of Ligand Binding in Structural Organization of Add A-riboswitch Aptamer: A Molecular Dynamics Simulation. J. Biomol. Struct. Dyn. 2011, 29, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Ríos, J. The THI-box Riboswitch, or How RNA Binds Thiamin Pyrophosphate. Structure 2007, 15, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Kubodera, T.; Watanabe, M.; Yoshiuchi, K.; Yamashita, N.; Nishimura, A.; Nakai, S.; Gomi, K.; Hanamoto, H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003, 555, 516–520. [Google Scholar] [CrossRef]
- Lu, C.; Smith, A.M.; Ding, F.; Chowdhury, A.; Henkin, T.M.; Ke, A. Variable sequences outside the SAM-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. J. Mol. Biol. 2011, 409, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Rieder, R.; Lang, K.; Graber, D.; Micura, R. Ligand-Induced Folding of the Adenosine Deaminase A-Riboswitch and Implications on Riboswitch Translational Control. ChemBioChem 2007, 8, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Lemay, J.-F.; Desnoyers, G.; Blouin, S.; Heppell, B.; Bastet, L.; St-Pierre, P.; Massé, E.; Lafontaine, D.A. Comparative Study between Transcriptionally- and Translationally-Acting Adenine Riboswitches Reveals Key Differences in Riboswitch Regulatory Mechanisms. PLoS Genet. 2011, 7, e1001278. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Yoon, J.; Hyeon, C.; Thirumalai, D. Using simulations and kinetic network models to reveal the dynamics and functions of riboswitches. Methods Enzymol. 2015, 553, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, A.J.; Lund, P.E.; Blanco, M.R.; Walter, N.G. The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts. Nat. Commun. 2016, 7, 8976. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Serganov, A.; Patel, D.J. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell 2010, 40, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Patel, D.J. Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol. 2012, 22, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Anthony, P.C.; Perez, C.F.; García-García, C.; Block, S.M. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc. Natl. Acad. Sci. USA 2012, 109, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-C.; Hyeon, C.; Thirumalai, D. Sequence-dependent folding landscapes of adenine riboswitch aptamers. Phys. Chem. Chem. Phys. 2014, 16, 6376–6382. [Google Scholar] [CrossRef] [PubMed]
- Montange, R.K.; Mondragón, E.; van Tyne, D.; Garst, A.D.; Ceres, P.; Batey, R.T. Discrimination between Closely Related Cellular Metabolites by the SAM-I Riboswitch. J. Mol. Biol. 2010, 396, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Priyakumar, U.D. Atomistic details of the ligand discrimination mechanism of S MK/SAM-III riboswitch. J. Phys. Chem. B 2010, 114, 9920–9925. [Google Scholar] [CrossRef] [PubMed]
- Doshi, U.; Kelley, J.M.; Hamelberg, D. Atomic-level insights into metabolite recognition and specificity of the SAM-II riboswitch. RNA 2012, 18, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Reiss, C.W.; Xiong, Y.; Strobel, S.A. Structural Basis for Ligand Binding to the Guanidine-I Riboswitch. Structure 2017, 25, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, J.; Lilley, D.M.J. The Structure of the Guanidine-II Riboswitch. Cell Chem. Biol. 2017, 24, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Sherlock, M.E.; Breaker, R.R. Biochemical Validation of a Third Guanidine Riboswitch Class in Bacteria. Biochemistry 2017, 56, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hwang, X.Y.; Stav, S.; Breaker, R.R. The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA 2016, 22, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Boyapati, V.K.; Huang, W.; Spedale, J.; Aboul-Ela, F. Basis for ligand discrimination between ON and OFF state riboswitch conformations: The case of the SAM-I riboswitch. RNA 2012, 18, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Liberman, J.A.; Wedekind, J.E. Riboswitch structure in the ligand-free state. Wiley Interdiscip. Rev. RNA 2012, 3, 369–384. [Google Scholar] [CrossRef] [PubMed]
- Allnér, O.; Nilsson, L.; Villa, A. Loop-loop interaction in an adenine-sensing riboswitch: A molecular dynamics study. RNA 2013, 19, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, S.; Prychyna, O.; Leung, J.; Avdic, D.; O’Neill, M.A. Ligand-directed dynamics of adenine riboswitch conformers. J. Am. Chem. Soc. 2007, 129, 11308–11309. [Google Scholar] [CrossRef] [PubMed]
- Kulshina, N.; Edwards, T.E.; Ferré-D’Amaré, A.R. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 2010, 16, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Tomšič, J.; McDaniel, B.A.; Grundy, F.J.; Henkin, T.M. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J. Bacteriol. 2008, 190, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.N.; Sosnick, T.R.; Pan, T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. USA 2007, 104, 17995–18000. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Bulusu, G.; Mitra, A. MD simulations of ligand-bound and ligand-free aptamer: Molecular level insights into the binding and switching mechanism of the add A-riboswitch. RNA 2009, 15, 1673–1692. [Google Scholar] [CrossRef] [PubMed]
- Rentmeister, A.; Mayer, G.; Kuhn, N.; Famulok, M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007, 35, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Kim, J.; Jha, S.; Aboul-ela, F. Conformational heterogeneity of the SAM-I riboswitch transcriptional ON state: A chaperone-like role for S-adenosyl methionine. J. Mol. Biol. 2012, 418, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Hennelly, S.P.; Sanbonmatsu, K.Y. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011, 39, 2416–2431. [Google Scholar] [CrossRef] [PubMed]
- Montange, R.K.; Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 2006, 441, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Lutz, B.; Faber, M.; Verma, A.; Klumpp, S.; Schug, A. Differences between cotranscriptional and free riboswitch folding. Nucleic Acids Res. 2014, 42, 2687–2696. [Google Scholar] [CrossRef] [PubMed]
- Lemay, J.-F.; Penedo, J.C.; Tremblay, R.; Lilley, D.M.J.; Lafontaine, D.A. Folding of the Adenine Riboswitch. Chem. Biol. 2006, 13, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Frieda, K.L.; Block, S.M. Direct Observation of Cotranscriptional Folding in an Adenine Riboswitch. Science 2012, 338, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, W.-B.; Chen, S.-J. Predicting Secondary Structural Folding Kinetics for Nucleic Acids. Biophys. J. 2010, 98, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Wang, Y.; Wang, Z.; Wang, Y.; Zhang, W. Reversible-Switch Mechanism of the SAM-III Riboswitch. J. Phys. Chem. B 2016, 120, 12305–12311. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Wang, Y.; Zhang, W. The regulation mechanism of yitJ and metF riboswitches. J. Chem. Phys. 2015, 143, 45103. [Google Scholar] [CrossRef] [PubMed]
- Kramer, F.R.; Mills, D.R. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981, 9, 5109–5124. [Google Scholar] [CrossRef] [PubMed]
- Proctor, J.R.; Meyer, I.M. CoFold: An RNA secondary structure prediction method that takes co-transcriptional folding into account. Nucleic Acids Res. 2013, 41, e102. [Google Scholar] [CrossRef] [PubMed]
- Heilman-Miller, S.L.; Woodson, S.A. Effect of transcription on folding of the Tetrahymena ribozyme. RNA 2003, 9, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Chadalavada, D.M.; Cerrone-Szakal, A.L.; Bevilacqua, P.C. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA 2007, 13, 2189–2201. [Google Scholar] [CrossRef] [PubMed]
- Mahen, E.M.; Watson, P.Y.; Cottrell, J.W.; Fedor, M.J. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010, 8, e1000307. [Google Scholar] [CrossRef] [PubMed]
- Diegelman-Parente, A.; Bevilacqua, P.C. A Mechanistic Framework for Co-transcriptional Folding of the HDV Genomic Ribozyme in the Presence of Downstream Sequence. J. Mol. Biol. 2002, 324, 1–16. [Google Scholar] [CrossRef]
- Danilova, L.V.; Pervouchine, D.D.; Favorov, A.V.; Mironov, A. a RNAKinetics: A web server that models secondary structure kinetics of an elongating RNA. J. Bioinform. Comput. Biol. 2006, 4, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Xayaphoummine, A.; Bucher, T.; Isambert, H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005, 33, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, W. Kinetic analysis of the effects of target structure on siRNA efficiency. J. Chem. Phys. 2012, 137, 225102. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gong, S.; Wang, Y.; Zhang, W. Kinetic partitioning mechanism of HDV ribozyme folding. J. Chem. Phys. 2014, 140, 25102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, W.; Chen, S.-J. Cotranscriptional folding kinetics of ribonucleic acid secondary structures. J. Chem. Phys. 2011, 135, 245101. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; SantaLucia, J.; Burkard, M.E.; Kierzek, R.; Schroeder, S.J.; Jiao, X.; Cox, C.; Turner, D.H. Thermodynamic Parameters for an Expanded Nearest-Neighbor Model for Formation of RNA Duplexes with Watson-Crick Base Pairs †. Biochemistry 1998, 37, 14719–14735. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.S.; von Hippel, P.H. Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 1995, 92, 8793–8797. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Svetlov, V.; Anthony, L.; Burgess, R.R.; Landick, R. RNA Polymerases from Bacillus subtilis and Escherichia coli Differ in Recognition of Regulatory Signals In Vitro. J. Bacteriol. 2000, 182, 6027–6035. [Google Scholar] [CrossRef] [PubMed]
- Mooney, R.A.; Artsimovitch, I.; Landick, R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 1998, 180, 3265–3275. [Google Scholar] [PubMed]
- Greenleaf, W.J.; Frieda, K.L.; Foster, D.A.N.; Woodside, M.T.; Block, S.M. Direct Observation of Hierarchical Folding in Single Riboswitch Aptamers. Science 2008, 319, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Monforte, J.A.; Kahn, J.D.; Hearst, J.E. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry 1990, 29, 7882–7890. [Google Scholar] [CrossRef] [PubMed]
- Komissarova, N.; Kashlev, M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. USA 1998, 95, 14699–14704. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 2000, 97, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Tolić-Nørrelykke, S.F.; Engh, A.M.; Landick, R.; Gelles, J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004, 279, 3292–3299. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Henkin, T.M. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998, 30, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Batey, R.T. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip. Rev. RNA 2011, 2, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, R.T.; Grundy, F.J.; Henkin, T.M. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc. Natl. Acad. Sci. USA 2007, 104, 4876–4880. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.T.; Moulin, M.; Webb, M.E.; Smith, A.G. Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl. Acad. Sci. USA 2007, 104, 20770–20775. [Google Scholar] [CrossRef] [PubMed]
- Haller, A.; Altman, R.B.; Souliere, M.F.; Blanchard, S.C.; Micura, R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc. Natl. Acad. Sci. USA 2013, 110, 4188–4193. [Google Scholar] [CrossRef] [PubMed]
- Cressina, E.; Chen, L.; Moulin, M.; Leeper, F.J.; Abell, C.; Smith, A.G. Identification of novel ligands for thiamine pyrophosphate (TPP) riboswitches. Biochem. Soc. Trans. 2011, 39, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Neupane, K.; Yu, H.; Foster, D.A.N.; Wang, F.; Woodside, M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011, 39, 7677–7687. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, S.; Wang, Y.; Wang, Z.; Zhang, W. Co-Transcriptional Folding and Regulation Mechanisms of Riboswitches. Molecules 2017, 22, 1169. https://doi.org/10.3390/molecules22071169
Gong S, Wang Y, Wang Z, Zhang W. Co-Transcriptional Folding and Regulation Mechanisms of Riboswitches. Molecules. 2017; 22(7):1169. https://doi.org/10.3390/molecules22071169
Chicago/Turabian StyleGong, Sha, Yanli Wang, Zhen Wang, and Wenbing Zhang. 2017. "Co-Transcriptional Folding and Regulation Mechanisms of Riboswitches" Molecules 22, no. 7: 1169. https://doi.org/10.3390/molecules22071169



