Possible Involvement of Hydrosulfide in B12-Dependent Methyl Group Transfer
Abstract
:1. Introduction and Background
2. Methylation, B12 Involvement, and Implication of Sulfur
3. The New Field of Sulfane Sulfur/Hydrogen Sulfide
4. New Knowledge on Cobalamin Hydrosulfide
5. Oxidation of Hydrosulfide Radical
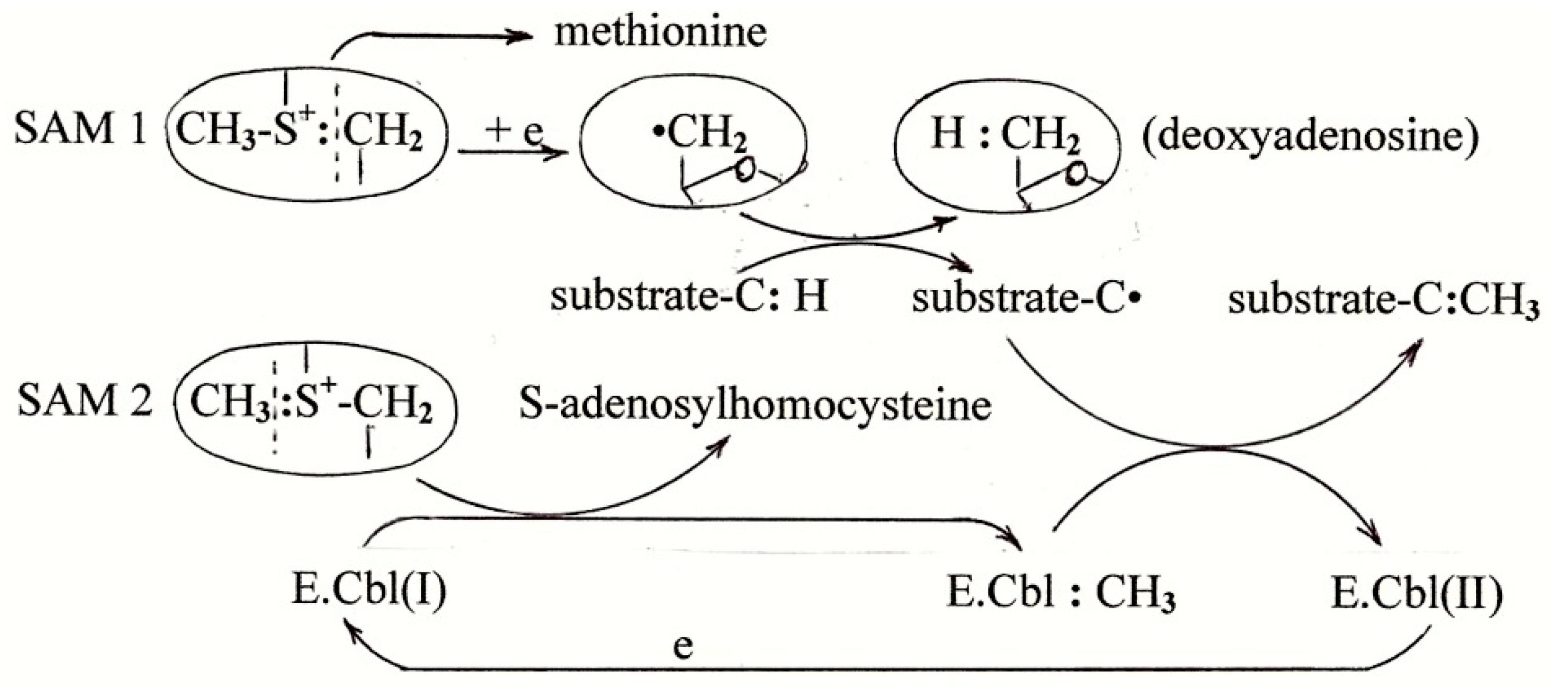
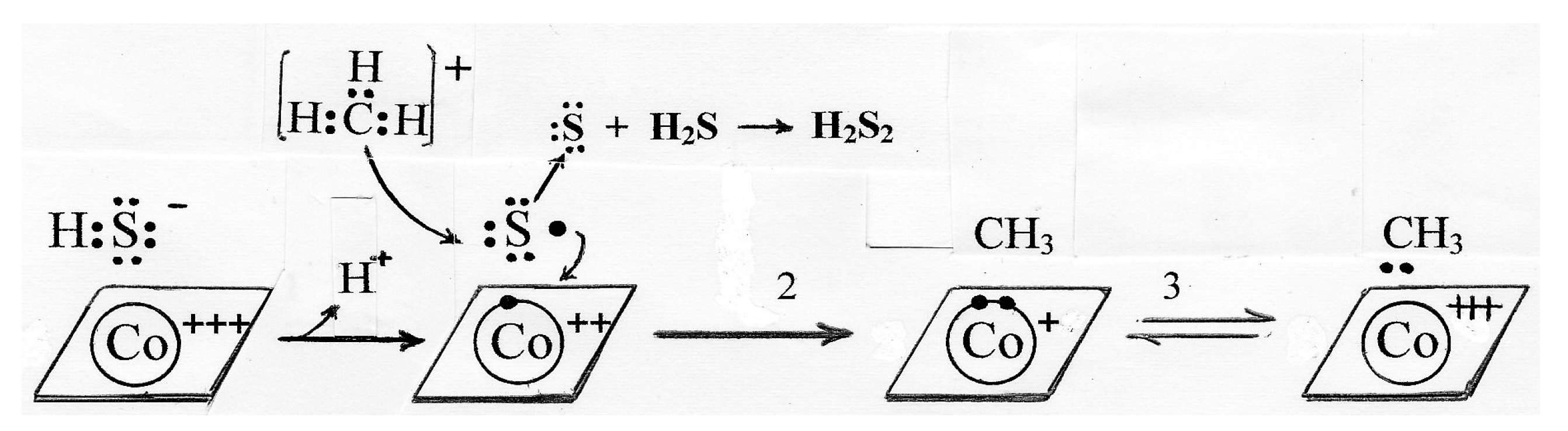
6. The Mechanism of B12-Dependent Radical SAM Methylation
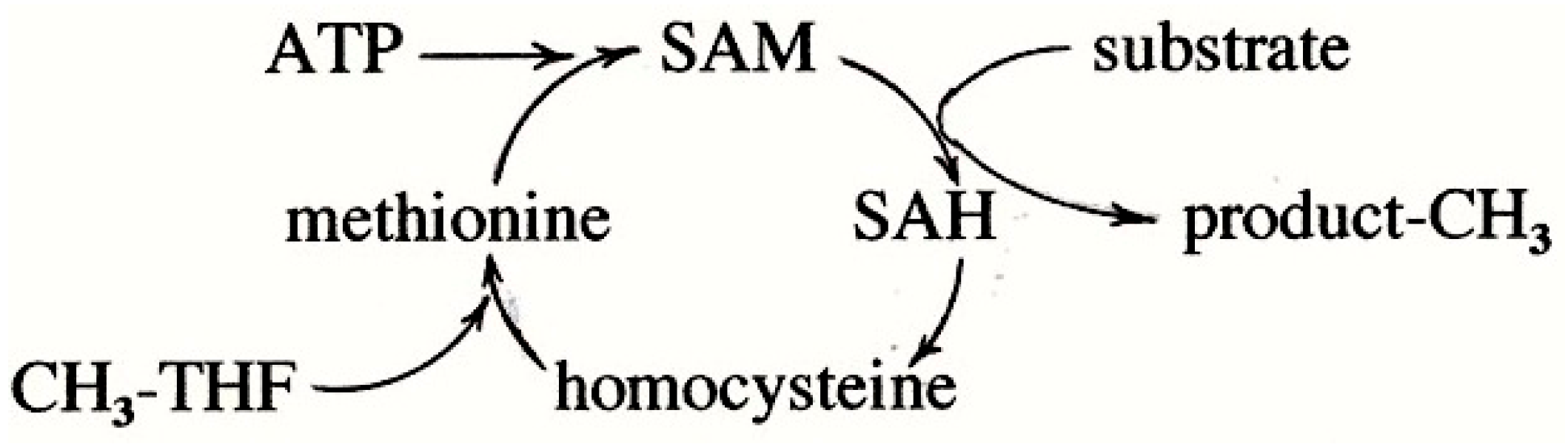
7. Methionine Synthesis
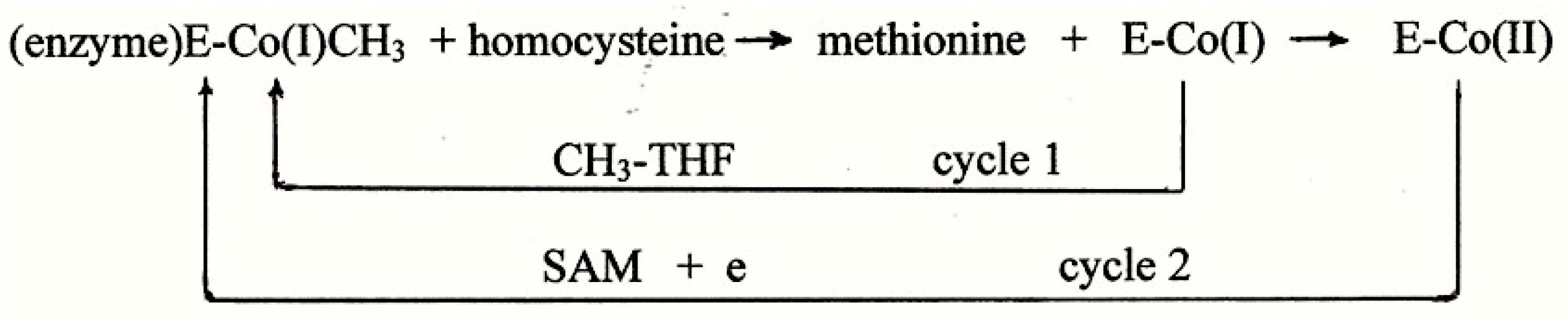
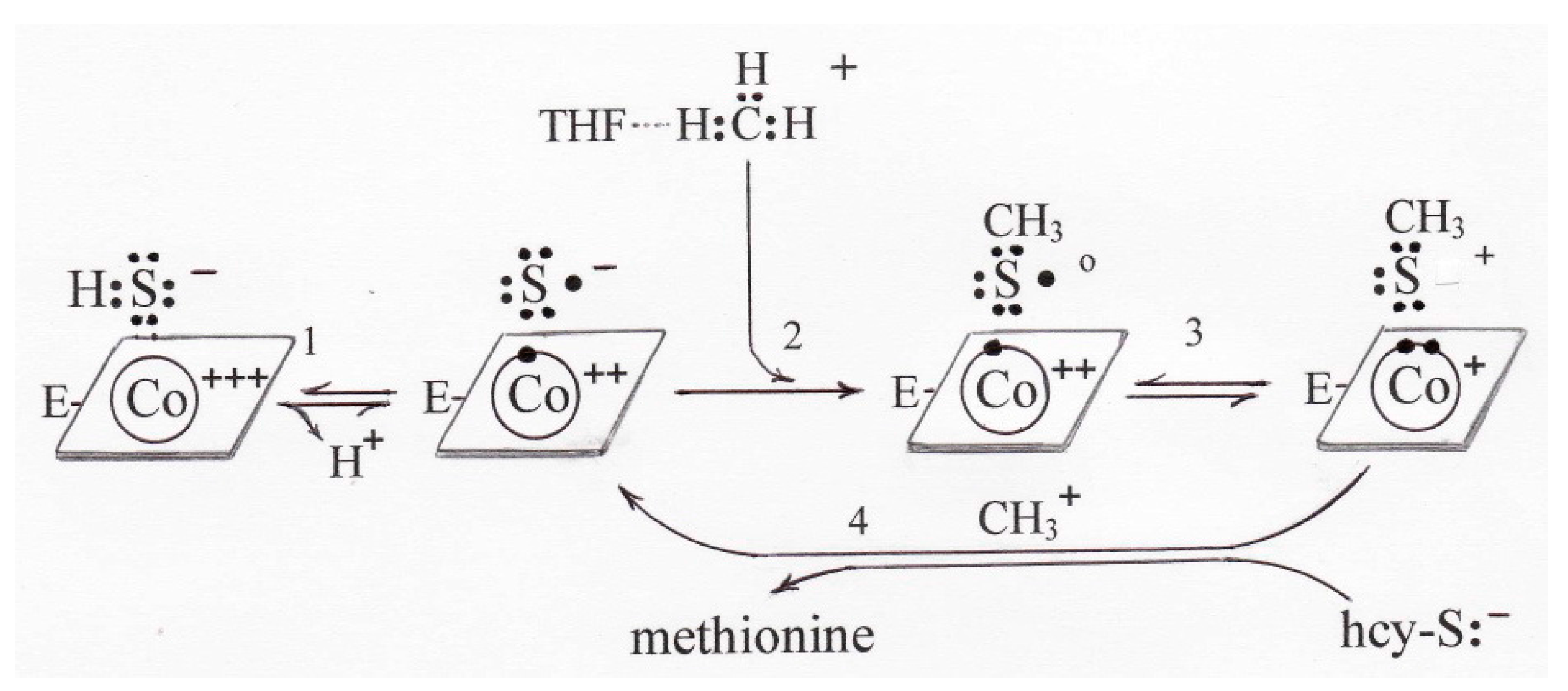
7.1. The Accepted Mechanism of Methionine Synthesis
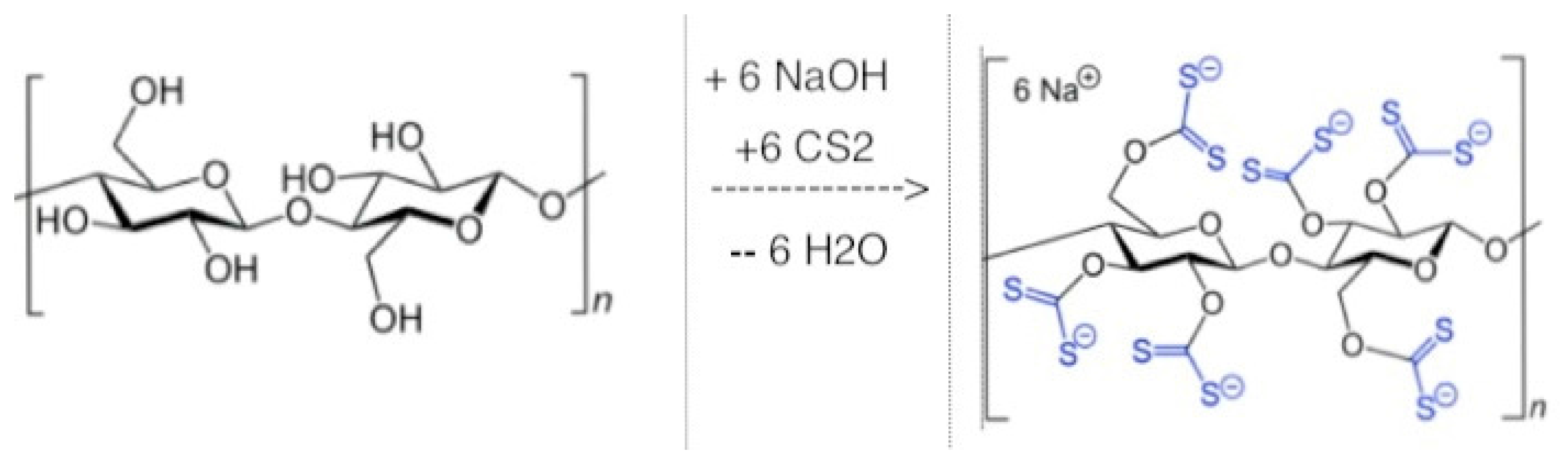
7.2. Viscose Dialysis Membrane
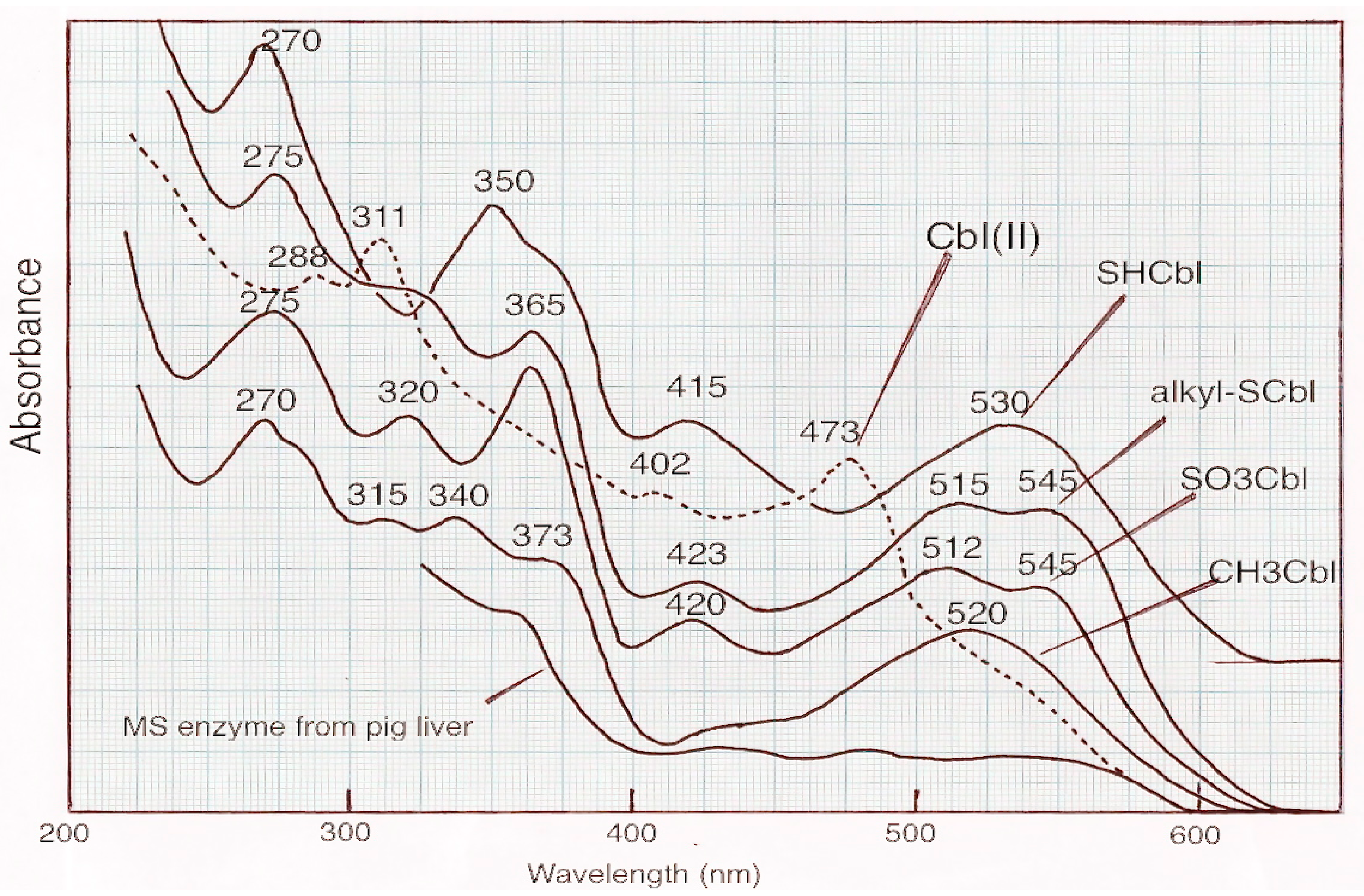
7.3. The Nature of the Cobalamin in Methionine Synthase
7.4. Excess Vitamin B12 Added to Cell Culture Greatly Increases MS Specific Activity
- (a)
- Animal cells grown in vitro secrete cysteine and homocysteine into the medium [29]
- (b)
- Cobalamin compounds are exceptionally effective in catalyzing the autoxidation of sulfhydryl compounds to disulfides: cys-SH + R-SH + ½O2 → cys-S-S-R + H2O [30]
- (c)
- Cysteine disulfides are degraded by C-S lyases to give persulfides according the following equation: cys-S-S-R → NH3 + pyruvate + cy-S-S-H [1].
7.5. Methionine Auxotrophy
8. Putting the Information Together
- (a)
- sulfur atom may facilitate the formation of MeCbl which acts as the methyl group carrier or
- (b)
- sulfur atom may bind to the cobalt and the cobalt-sulfur unit acts as the methyl group carrier.
8.1. Possible Catalytic Role of Sulfur in Formation of MeCbl
8.2. Sulfur as Part of the Carrier in Methionine Synthase
- -
- crude tissue extracts which have not been manipulated in such a way that the sulfur is lost,
- -
- purified enzyme which has been protected by thiols during purification,
- -
- purified enzyme that has been exposed for prolonged periods to dialysis membrane, and
- -
- crude extracts of cells which have been cultured in the presence of ~1 μM vitamin B12 under conditions that can generate S° (as in reference [28]).
8.3. Sulfur as Part of the Carrier in RSMT
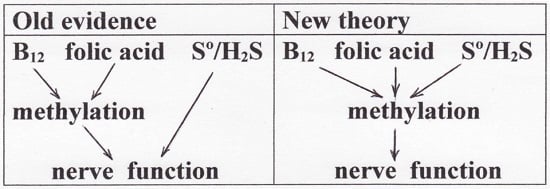
8.4. The Hypomethylation Theory and Dementia
9. Conclusions and Perspective
- −
- isolation of sulfitoCbl from the air-exposed purified enzyme,
- −
- decreased need for SAM in much-dialyzed enzyme (given that viscose membrane releases S°),
- −
- 30-fold increase in activity in cells cultured in conditions which favor S° formation, and
- −
- methionine auxotrophy in cells with defective sulfur metabolism.
Conflicts of Interest
References
- Toohey, J.I.; Cooper, A.J.L. Thiosulfoxide (sulfane) sulfur: New chemistry and new roles in biology. Molecules 2014, 19, 12789–12813. [Google Scholar] [CrossRef] [PubMed]
- Iciek, M.; Kowalszyzk-Pachel, D.; Biska-Wilkosz, A.; Gorny, M.; Wlodek, L. S-sulfhydration as a cellular redox regulation. Biosci. Rep. 2015, 36, e00304. [Google Scholar] [CrossRef] [PubMed]
- Kasamatsu, S.; Nishimura, A.; Morita, M.; Matsunaga, T.; Hamid, H.A.; Akaike, T. Redox signaling regulated by cysteine persufide and protein polysulfidation. Molecules 2016, 21, 1721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; van der Donk, W.A.; Liu, W. Radical-mediated enzymatic methylation: A tale of two SAMs. Acc. Chem. Res. 2012, 45, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Wing, D.; Li, A.; Jia, Y.; Ji, X.; Qianzhu, H.; Zhang, Q. Emerging diversity of the cobalamin-dependent methyltransferases involving radical-based mechanisms. ChemBioChem 2016, 17, 1191–1197. [Google Scholar]
- Matthews, R.G. Methionine biosynthesis. In Methionine Biosynthesis in Folates and Pterins; Blakely, R.L., Ed.; John Wiley & Sons: New York, NY, USA, 1984; Volume 1, pp. 497–550. [Google Scholar]
- Stupperich, E. Recent advances in elucidation of biological corrinoid functions. FEMS Microbiol. Rev. 1993, 12, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Toohey, J.I. Sulfur signaling: Is the agent sulfide or sulfane. Anal. Biochem. 2011, 413, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ida, T.; Sawa, T.; Ihari, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fjii, S.; Matsunaga, T.; et al. Reactive cysteine persulfide and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [PubMed]
- Toohey, J.I. Persulfide sulfur is a growth factor for cells defective in sulfur metabolism. Biochem. Cell Biol. 1986, 4, 758–765. [Google Scholar] [CrossRef]
- Toohey, J.I. Hydrosulfide derivatives of cobalamins. J. Inorg. Biochem. 1993, 49, 189–199. [Google Scholar] [CrossRef]
- Salnikov, D.S.; Kucherenko, P.N.; Dereven’kov, I.A.; Makarov, S.V.; van Eldik, R. Kinetics and mechanism of the reaction of hydrogen sulfide with cobalamin in aqueous solution. Eur. J. Inorg. Chem. 2014, 2014, 852–862. [Google Scholar] [CrossRef]
- Salnikov, D.S.; Makarov, S.V.; Eldik, R.; Kucherenko, P.N.; Boss, G.R. Kinetics and mechanism of the reaction hydrogen sulfide with diaquacobinamide in aqueous solution. Eur. J. Inorg. Chem. 2014, 2014, 4123–4133. [Google Scholar] [CrossRef] [PubMed]
- Strianese, M.; Mirra, S.; Bertolasi, V.; Milione, S.; Pellecchia, C. Organometallic sulfur complexes: Reactivity of the hydrogen sulfide anion with cobaloximes. New J. Chem. 2015, 39, 4093–4099. [Google Scholar] [CrossRef]
- Adler, N.; Medwick, T.; Poznanski, T.J. Reaction of hydroxocobalamin with thiols. J. Am. Chem. Soc. 1966, 88, 5018–5020. [Google Scholar] [CrossRef]
- Zhu, J.; Petit, K.; Colson, A.O.; deBolt, S.; Sevilla, M.D. Reactions of sulfhydryl and sulfide radicals with oxygen, hydrogen sulfide, hydrosulfide, and sulfide: Formation of SO2−, HSSH−, HSS.2− and HSS. J. Phys. Chem. 1991, 95, 3676–3681. [Google Scholar] [CrossRef]
- Chen, Z.; Crippen, K.; Gulati, S.; Banerjee, R. Purification and kinetic mechanism of a mammalian methionine synthase from pig liver. J. Biol. Chem. 1994, 269, 27193–27197. [Google Scholar] [PubMed]
- Loughlin, R.E.; Elford, H.L.; Buchanan, J.M. Enzymatic synthesis of the methyl group of methionine: VII Isolation of a cobalamin-containing transmethylase (5′-methyltetrahydrofolate-homocysteine) from mammalian liver. J. Biol. Chem. 1964, 239, 2888–2895. [Google Scholar] [PubMed]
- Mangum, J.H.; North, J.A. Isolation of a cobalamin containing 5-methyltetrahydrofolate-homocysteine transmethylase from mammalian liver. Biochemistry 1971, 10, 3765–3769. [Google Scholar] [CrossRef] [PubMed]
- Rudiger, H.; Jaenicke, L. Methionine synthase: Existence and interconversion of two enzyme species. Eur. J. Biochem. 1970, 16, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Toohey, J.I.; (University of California, Los Angeles, CA, USA). Unpublished work. 1980.
- Xanthogenate Cellulose Structural Formula V1.svg. Available online: https://commons.wikimedia.org/wiki/File:Xanthogenate_Cellulose_Structural_Formula_V1.svg#file (accessed on 5 February 2017).
- You, Z.; Cao, X.; Taylor, A.B.; Hart, P.J.; Levine, R. Characterization of a covalent polysulfane bridge in Cu-Zn superoxide dismutase. Biochemistry 2010, 49, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Fuji, K.; Huennekens, F.M. Activation of methionine synthase by a reduced triphosphopyridine nucleotide-dependent flavoprotein system. J. Biol. Chem. 1974, 249, 6745–6753. [Google Scholar]
- Taylor, R.T.; Weissbach, H. N5-Methyltetrahydrofolate-homocysteine transmethylase: Partial purification and properties. J. Biol. Chem. 1967, 242, 1502–1508. [Google Scholar]
- Takeyama, S.; Buchanan, J.M. Enzymatic synthesis of the methyl group of methionine: III Spectral and electrophoretic studies of the prosthetic group of the B12 enzyme. J. Biochem. 1961, 49, 578–588. [Google Scholar] [CrossRef]
- Ertel, R.; Brot, N.; Taylor, R.; Weissbach, H. Studies on the nature of the bound cobamide in E. coli N5-methyltetrahydrofolate-homocysteine transmethylase. Arch. Biochem. Biophys. 1968, 126, 353–357. [Google Scholar] [CrossRef]
- Mangum, J.H.; Murray, B.K.; North, J.A. Vitamin B12 dependent methionine biosynthesis in cultured mammalian cells. Biochemistry 1969, 8, 3496–3499. [Google Scholar] [CrossRef] [PubMed]
- Bannai, S.; Ishii, T. Formation of sulfhydryl groups in the culture medium of human diploid fibroblasts. J. Cell. Physiol. 1980, 104, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, D.W.; Troxell, L.S.; Brown, K.L. Catalysis of thiol oxidation by cobalamins and cobinamides: Reaction products and kinetics. Biochemistry 1984, 23, 2017–2025. [Google Scholar] [CrossRef]
- Halpern, B.C.; Clark, B.R.; Hardy, D.N.; Halpern, R.M.; Smith, R.A. The effect of replacement of methionine by homocysteine on survival of malignant and normal adult mammalian cell in culture. Proc. Natl. Acad. Sci. USA 1974, 71, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Cellarier, E.; Durando, X.; Vasson, M.P.; Farges, M.C.; Demiden, A.; Maurizis, J.C.; Madelmont, J.C.; Chollet, P. Methionine dependency and cancer treatment. Cancer Treat. Rev. 2003, 29, 489–499. [Google Scholar] [CrossRef]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span expansion. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Watkins, D. Cobalamin metabolism in methionine-dependent human tumour and leukemia cell lines. Clin. Investig. Med. 1998, 21, 151–158. [Google Scholar]
- Loewy, A.D.; Niles, K.M.; Anastasio, N.; Watkins, D.; Lavoie, J.; Lerner-Ellis, J.P.; Pastinen, T.; Trasler, J.M.; Rosenblatt, D.S. Epigenetic modification of the gene for the vitamin B12 chaperone MMACHC can results in increased tumorigenicity and methionine dependence. Mol. Genet. Metab. 2009, 96, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Chu, R.C.; Begley, J.A.; Colligan, P.D.; Hall, C.A. The methylcobalamin metabolism of cultured human fibroblasts. Metabolism 1993, 42, 315–319. [Google Scholar] [CrossRef]
- Hannibal, L.; Kim, J.; Brasch, N.E.; Wang, S.; Rosenblatt, D.S.; Banerjee, R.; Jacobsen, D.W. Processing of alkylcobalamins in mammalian cells: A role of MMACHC (cblC) gene product. Mol. Genet. Metab. 2009, 97, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Hogenkamp, H.P.; Bratt, G.T.; Sun, S.Z. Methyl transfer from methylcobalamin: A reinvestigation. Biochemistry 1985, 24, 6428–6432. [Google Scholar] [CrossRef] [PubMed]
- Dolphin, D. Preparation of the reduced forms of Vitamin B12 and of some analogs of the Vitamin B12 coenzyme containing a cobalt-carbon bond. Meth. Enzymol. 1971, 18C, 34–52. [Google Scholar]
- Dolphin, D.H.; Johnson, A.W. The reaction of cobalamins with thiols: An alternative synthesis of alkyl-cobamide coenzyme analogues. Chem. Soc. Proc. 1963, 311–312. [Google Scholar]
- Taylor, R.T.; Hanna, M.L. Spectrophotometric evidence for the formation of an Eschericia coli B12s methyltransferase. Biochem. Biophys. Res. Commun. 1970, 38, 758–763. [Google Scholar] [CrossRef]
- Benjdia, A.; Pierre, S.; Gherasim, C.; Guillot, A.; Carmona, M.; Amara, P.; Banerjee, R.; Berteau, O. The thiostrepton A tryptophan methyltransferase TsrM catalyses a Cob(II)alamin-dependent methyl transfer reaction. Nat. Commun. 2015, 6, 8377. [Google Scholar] [CrossRef] [PubMed]
- Weir, D.G.; Scott, J.M. The biochemical basis of the neuropathy in cobalamin deficiency. Bailliere’s Clin. Haematol. 1995, 8, 479–497. [Google Scholar] [CrossRef]
- Reynolds, E.H. The neurology of folic acid deficiency. Handb. Clin. Neurol. 2014, 120, 927–943. [Google Scholar] [PubMed]
- Spencer, J.D. Metabolic vitamin B12 deficiency: A missed opportunity to prevent dementia and stroke. Nutr. Res. 2016, 36, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, H.; Yu, M.; Zhang, Y.; Liu, H.; Wilson, J.X.; Huang, G. Folic acid alters methylation profile of JAK-STAT and long-term depression signaling pathways in Alzheimer’s disease models. Mol. Neurobiol. 2016, 53, 6548–6556. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.E.; Sepchry, A.A.; Wangsgaard, J.D.; Koenig, J.E. The effect of S-adenosylmethionine on cognitive performance in mice; an animal model meta analysis. PLoS ONE 2014, 9, e107756. [Google Scholar] [CrossRef] [PubMed]
- Hyland, K.; Smith, J.; Bottiglieri, T.; Perry, J.; Wendel, U.; Clayton, P.T.; Leonard, J.V. Demyelination and decreased S-adenosylmethionine in 5,10-methylenetetrahydrofolate reductase deficiency. Neurology 1988, 38, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, M.E.; Ubagai, T.; Mudd, S.H.; Wilson, W.G.; Leonard, J.V. Demyelination of the brain is associated with methionine adenosyltransferase deficiency. J. Clin. Investig. 1996, 98, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.H.; Shi, X.R.; Hong, Z.Y.; Pan, L.L.; Liu, X.H.; Zhu, Y.Z. A new hope for neurodegeneration: Possible role of hydrogen sulfide. J. Alzheimers Dis. 2011, 24, 13–182. [Google Scholar]
- Wei, H.J.; Li, X.; Tang, X.Q. Therapeutic benefits of H2S in Alzheimers disease. J. Clin. Neurosci. 2014, 21, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, P. Endogenous production of hydrogen sulfide in mammals. Amino Acids 2004, 26, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H.; Beck, P.W. Characterization of the enyzmic capacity for cysteine desulphhydration in liver and kidney of the rat. Biochem. J. 1982, 206, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Guiraud, S.P.; Montoliu, L.; de Silva, L.; Dayon, L.; Galindo, A.N.; Martin, J.C. High throughput and simultaneous quantitative analysis of homocysteine-methionine cycle metabolites and cofactors in blood plasma and cerebrospinal fluid by isotope dilution LS-MS/MS. Anal. Bioanal. Chem. 2017, 409, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, N.; Tanaka, M.; Yoshida, M.; Ogasawara, Y.; Togawa, T.; Ishii, K.; Kimura, H. 3-Mercaptopyruvate sulfrtransferase produces hydrogen sulfide and bound sulfur in the brain. Antioxid. Redox Signal. 2009, 11, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, R.; Otsuguro, K.; Yamaguchi, S.; Ito, S. Contribution of cysteine aminotransferase and mercaptopyruvate sulfurtransferase to hydrogen sulfide produciton in peripheral neurons. J. Neurochem. 2014, 130, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, N.; Nagano, M.; Ito, T.; Shinamura, K.; Akimoto, T.; Suzuki, H. Antioxidant enzyme, 3-mercaptopyruvate sulfurtransferase knockout mice exhibit increased anxiety-like behaviors: A moldel for human mercaptolactate-cysteine disulfidurea. Sci. Rep. 2013, 3, 1986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.L.; Fang, F.; Qiao, P.F.; Yan, N.; Gao, D.; Yan, Y. AP39, a mitochondria-targeted sulfide donor, supports cellular bioenergetics and protects against Alzheimer’s disease by preserving mitochondrial funcion in APP/PS1 mice and neurons. Oxid. Med. Cell. Longev. 2016, 2016, 8360738. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

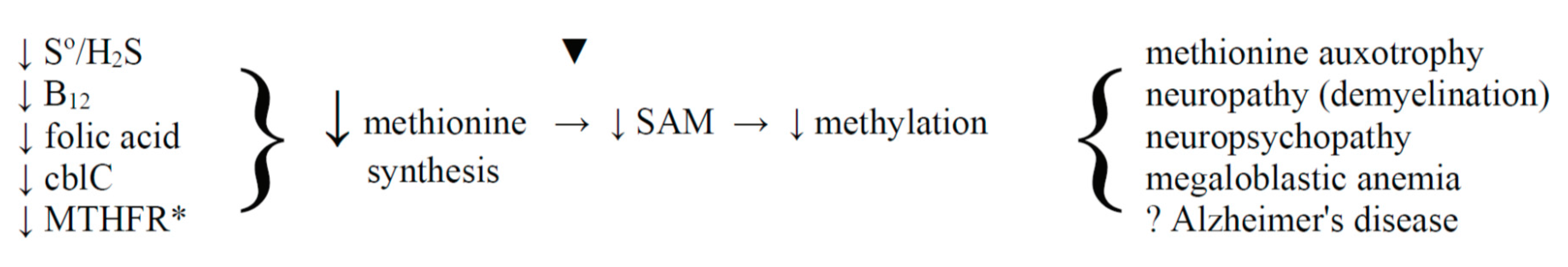
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toohey, J.I. Possible Involvement of Hydrosulfide in B12-Dependent Methyl Group Transfer. Molecules 2017, 22, 582. https://doi.org/10.3390/molecules22040582
Toohey JI. Possible Involvement of Hydrosulfide in B12-Dependent Methyl Group Transfer. Molecules. 2017; 22(4):582. https://doi.org/10.3390/molecules22040582
Chicago/Turabian StyleToohey, John I. 2017. "Possible Involvement of Hydrosulfide in B12-Dependent Methyl Group Transfer" Molecules 22, no. 4: 582. https://doi.org/10.3390/molecules22040582