Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay
Abstract
:1. Introduction
2. Results
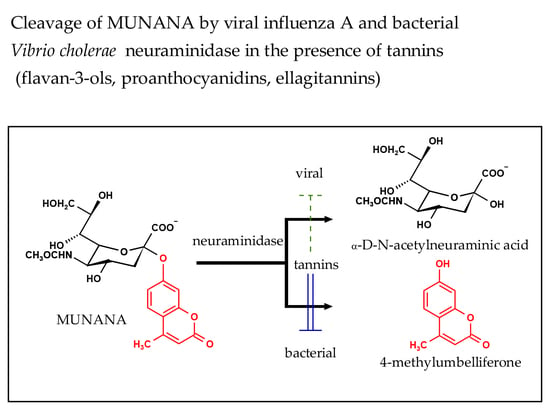
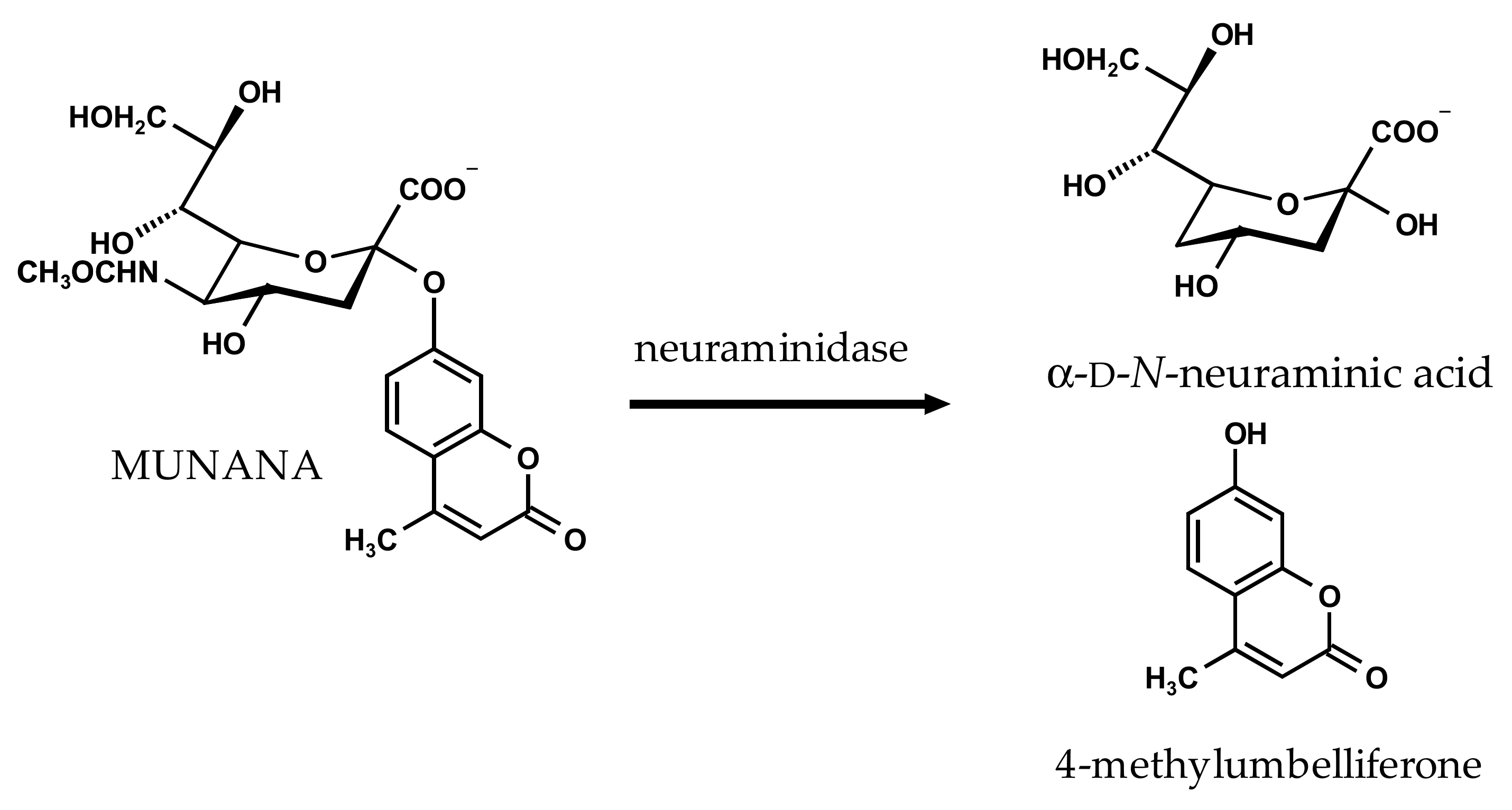
2.1. Inhibition of Viral Influenza A Neuraminidase and Bacterial Vibrio cholerae Neuraminidase by the Reference Compounds
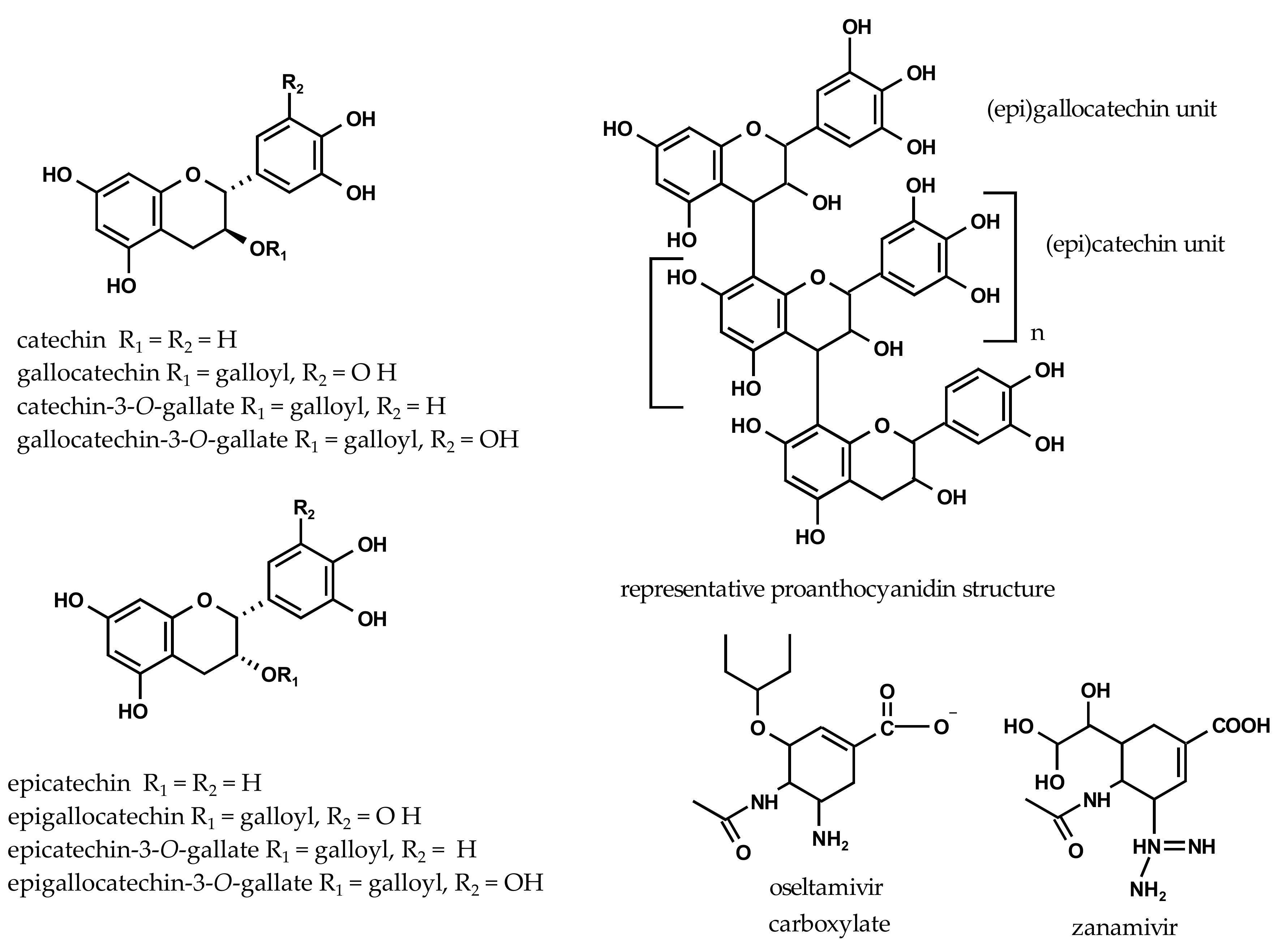
2.2. Inhibition of Viral H1N1-NA and Bacterial VCNA by Flavan-3-ols
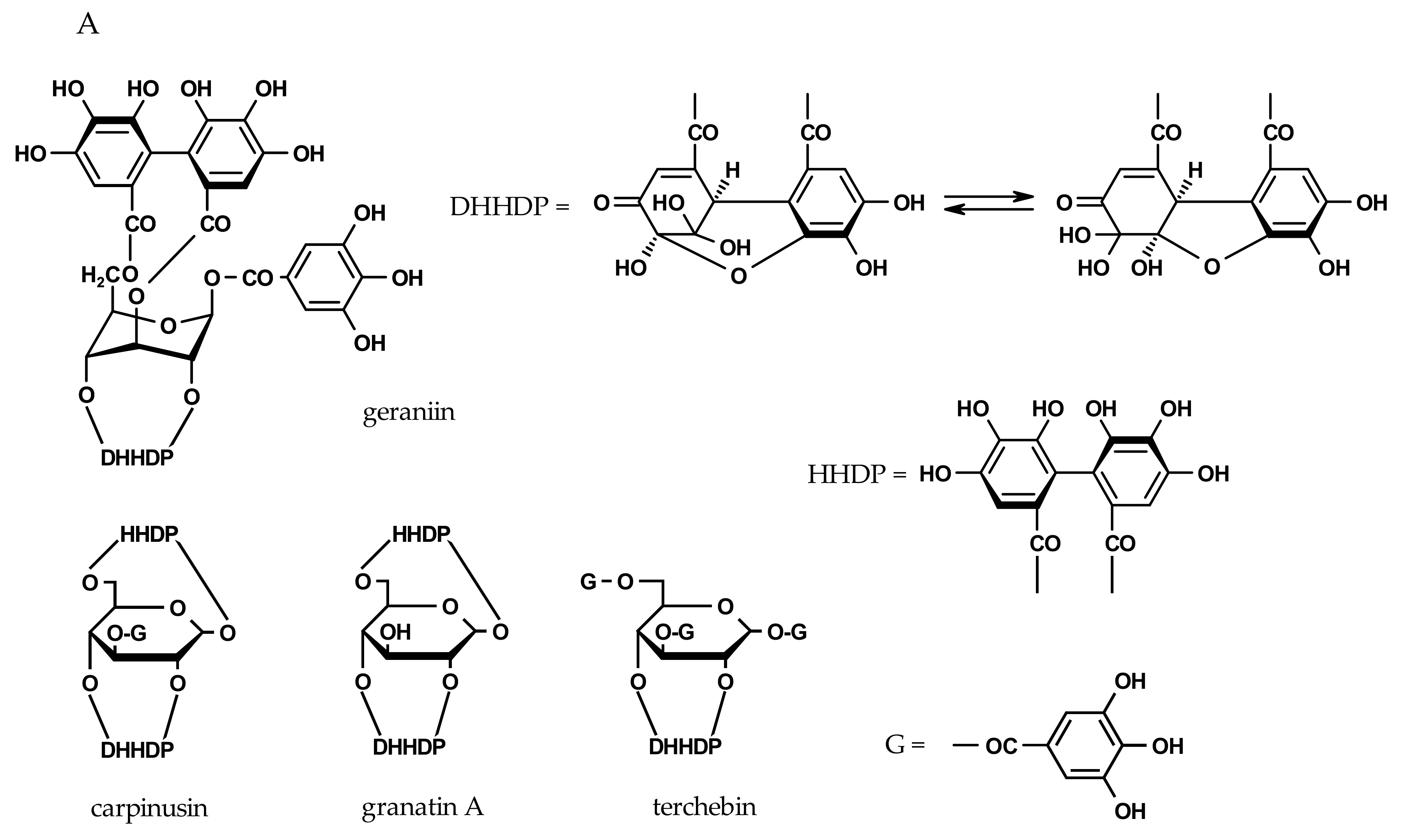
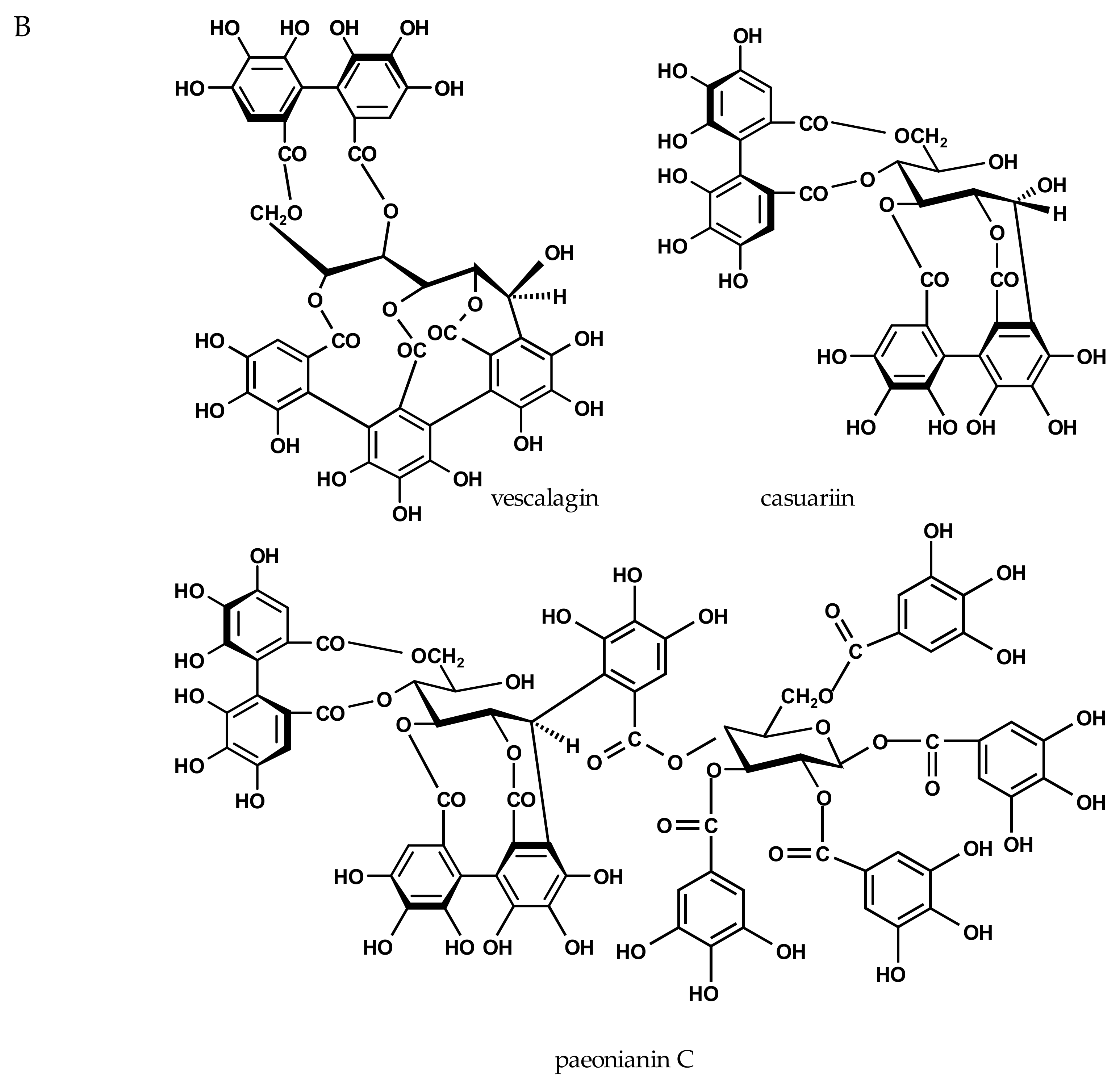
2.3. Inhibition of Viral H1N1-NA and Bacterial VCNA by Ellagitannins
2.4. Inhibition of Viral H1N1-NA and Bacterial VCNA by Plant-Derived Fractions
2.5. Combined Effect of Zanamivir and EPs® 7630 on VCNA Activity
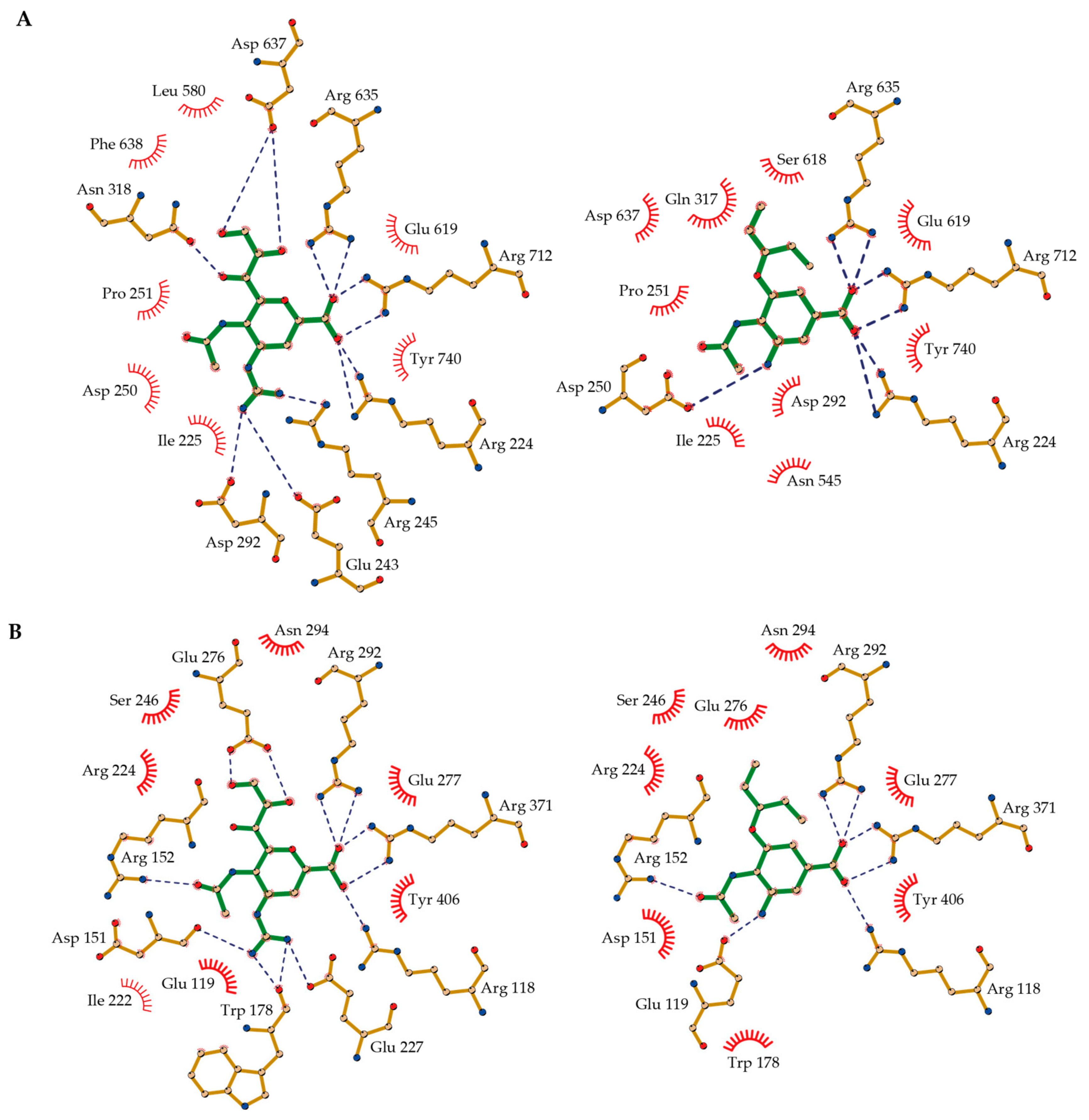
2.6. Molecular Basis of NA-Inhibitor Interactions as Assessed by Crystallographic Analyses
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. NA Inhibition Assay
4.3. Drug Combination Analysis
4.4. Expression and Purification of Recombinant VCNA
4.5. VCNA Crystallization and Structure Determination
4.6. Multiple Sequence Alignment
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Fry, A.M.; Gubareva, L.V. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir. Ther. 2012, 17, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Grienke, U.; Schmidtke, M.; von Grafenstein, S.; Kirchmair, J.; Liedl, K.R.; Rollinger, J.M. Influenza neuraminidase: A druggable target for natural products. Nat. Prod. Rep. 2012, 29, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Rakers, C.; Schwerdtfeger, S.M.; Mortier, J.; Duwe, S.; Wolff, T.; Wolber, G.; Melzig, M.F. Inhibitory potency of flavonoid derivatives on influenza virus neuraminidase. Bioorg. Med. Chem. Lett. 2014, 24, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Lee, K.H.; Seong, B.L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005, 68, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jia, W.; Zhao, A. Anti-influenza agents from plants and traditional Chinese medicine. Phytother. Res. 2006, 20, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Wang, Y.F.; Xu, J.; Gu, Q.; Liu, H.B.; Xiao, P.G.; Zhou, J.; Liu, Y.; Yang, Z.; Su, H. Anti-influenza agents from traditional Chinese medicine. Nat. Prod. Rep. 2010, 27, 1758–1780. [Google Scholar] [CrossRef] [PubMed]
- Droebner, K.; Ehrhardt, C.; Poetter, A.; Ludwig, S.; Planz, O. CYSTUS052, a polyphenol-rich plant extract, exerts anti-influenza virus activity in mice. Antivir. Res. 2007, 76, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Hrincius, E.R.; Korte, V.; Muzur, I.; Droebner, K.; Poetter, A.; Dresschers, S.; Schmolke, M.; Planz, O.; Ludwig, S. A polyphenol-rich plant extract, CYSTUS=52, exerts anti influenza virus activity in cell culture without toxic side effects or the tendency to induce viral resistance. Antivir. Res. 2007, 76, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Schwerdtfeger, S.M.; Melzig, M.F. Sialidases in biological systems. Pharmazie 2010, 65, 551–561. [Google Scholar] [PubMed]
- Kelly, R.T.; Farmer, S.; Greiff, D. Neuraminidase activities of clinical isolates of Diplococcus pneumoniae. J. Bacteriol. 1967, 84, 272–273. [Google Scholar]
- Pettigrew, M.M.; Fennie, K.P.; York, M.P.; Daniels, J.; Ghaffar, F. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect. Immun. 2006, 74, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.L.; Lewis, W.G. Host sialoglycans and bacterial sialidases: A mucosal perspective. Cell. Microbiol. 2012, 14, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Soong, G.; Muir, A.; Gomez, M.I.; Waks, J.; Reddy, B.; Planet, P.; Singh, P.K.; Kaneko, Y.; Wolfgang, M.C.; Hsiao, Y.S.; et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Investig. 2006, 116, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A.; Bartmess, K.C. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 2003, 187, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.C.; Thomson, R.J.; Dyason, J.C.; McAtamney, S.; von Itzstein, M. Modelling, synthesis and biological evaluation of novel glucuronide-based probes of Vibrio cholerae sialidase. Med. Chem. 2006, 14, 1518–1537. [Google Scholar] [CrossRef] [PubMed]
- Potier, M.; Mameli, L.; Belisle, M.; Dallaire, L.; Melancon, S. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal. Biochem. 1979, 94, 287–296. [Google Scholar] [CrossRef]
- Quosdorf, S.J.; Kolodziej, H. The potential of flavan-3-ols and their oligomers as neuraminidase inhibitors and limitation of a fluorescence-based assay. In Proceedings of the 8th ISANH Congress on Polyphenols Applications, Lisbon, Portugal, 5–6 June 2014; p. 237. [Google Scholar]
- Vavricka, C.J.; Li, Q.; Wu, Y.; Qi, J.; Wang, M.; Liu, Y.; Gao, F.; Liu, J.; Feng, E.; He, J. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog. 2011, 7, e1002249. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Shimizu, K.; Tanaka, T.; Kuroda, K.; Takayama, T.; Yamamoto, T.; Hanada, N.; Hamada, Y. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS ONE 2012, 7, e45371. [Google Scholar] [CrossRef] [PubMed]
- Govorkova, E.A.; Fang, H.B.; Tan, M.; Webster, R.G. Neuraminidase inhibitor-rimantadine combinations exert additive and synergistic anti-influenza virus effects in MDCK Cells. Antimicrob. Agents Chemother. 2004, 48, 4855–4863. [Google Scholar] [CrossRef] [PubMed]
- Ilyushina, N.A.; Bovin, N.V.; Webster, R.S.; Govorkan, E.A. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antivir. Res. 2006, 70, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Ilyushina, N.A.; Hoffmann, E.; Salomon, R.; Webster, R.S.; Govorkan, E.A. Amantadine-oseltamivir combination therapy for H5N1 influenza virus infection in mice. Antivir. Ther. 2007, 12, 363–370. [Google Scholar] [PubMed]
- Ilyushina, N.A.; Hay, A.; Yilmaz, N.; Boon, A.C.; Webster, R.S.; Govorkan, E.A. Oseltamivir-ribavirin combination therapy for highly pathogenic H5N1 influenza virus infection in mice. Antimicrob. Agents Chemother. 2008, 52, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Charyasriwong, S.; Watanabe, K.; Rahmasari, R.; Matsunaga, A.; Haruyama, T.; Kobayashi, N. In vitro evaluation of synergistic inhibitory effects of neuraminidase inhibitors and methylglyoxal against influenza virus infection. Arch. Med. Res. 2015, 46, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Haidari, M.; Ali, M.; Casscells, S.W.; Madjid, M. Pomegranate (Punica granatum) purified extracts inhibits influenza virus and has a synergistic effect with oseltamivir. Phytomedicine 2009, 16, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Chou, T. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Kolodziej, H. Fascinating metabolic pools of Pelargonium sidoides and Pelargonium reniforme, traditional and phytomedicinal sources of the herbal medicine Umckaloabo®. Phytomedicine 2007, 14 (Suppl. VI), 18–26. [Google Scholar] [CrossRef] [PubMed]
- Bijnsdorp, I.V.; Giovannetti, E.; Peters, G.J. Analysis of drug interactions. Methods Mol. Biol. 2011, 731, 421–434. [Google Scholar] [PubMed]
- Schoetz, K.; Erdelmeier, C.; Germer, S.; Hauer, H. A detailed view on the constituents of EPs® 7630. Planta Med. 2008, 74, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Lawer, W.; Coleman, P.M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 1983, 303, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Colman, P.M.; Varghese, J.N.; Laver, W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983, 303, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Crennell, S.; Garman, E.; Laver, G.; Vimr, E.; Taylor, G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 1994, 2, 535–544. [Google Scholar] [CrossRef]
- Taylor, G. Sialidases: Structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996, 6, 830–837. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, MB. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.L.; Wang, H.D.; Lee, S.M.Y.; Wang, Y.T.; Du, G.H. Structure–activity relationships of flavonoids as influenza virus neuraminidase inhibitors and their in vitro antiviral activities. Bioorg. Med. Chem. 2008, 16, 7141–7147. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Tendulkar, A.V.; Wangikar, P.P. Drug discovery against H1N1 virus (influenza virus) via computational virtual screening approach. Med. Chem. Res. 2011, 20, 1445–1449. [Google Scholar] [CrossRef]
- Kolodziej, H. Antimicrobial, antiviral and immunomodulatory activity studies of Pelargonoium sidoides (EPs® 7630) in the context of health promotion. Pharmaceuticals 2011, 4, 1295–1314. [Google Scholar] [CrossRef] [PubMed]
- Juge, N.; Tailford, L.; Owen, C.D. Sialidases from gut bacteria: A mini-review. Biochem. Soc. Trans. 2016, 44, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Gut, H.; Xu, G.; Taylor, G.L.; Walsh, M.A. Structural basis for Streptococcus pneumoniae NanA inhibition by influenza antivirals zanamivir and oseltamivir carboxylate. J. Mol. Biol. 2011, 408, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Owen, C.D.; Walshaw, J.; Crost, E.H.; Hardy-Goddard, J.; le Gall, G.; de Vos, W.M.; Taylor, G.L.; Juge, N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat. Commun. 2015, 6, 7624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, C.D.; Lukacik, P.; Potter, J.A.; Sleator, O.; Taylor, G.L.; Walsh, M.A. Streptococcus pneumoniae NanC—Structural insights into the specificity and mechanism of a sialidase that produces a sialidase inhibitor. J. Biol. Chem. 2015, 290, 27736–27748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Schumann, L.; Walther, E.; Hoffmann, A.; Braun, H.; Grienke, U.; Rollinger, J.M.; von Grafenstein, S.; Liedl, K.R.; Kirchmair, J.; et al. Complementary assays helping to overcome challenges for identifying neuraminidase inhibitors. Future Virol. 2015, 10, 77–88. [Google Scholar] [CrossRef]
- Kolodziej, H. Ocurrence of procyanidins in Nelia meyeri. Phytochemistry 1984, 23, 1745–1752. [Google Scholar] [CrossRef]
- Kolodziej, H. Oligomeric flavan-3-ols from medicinal willow bark. Phytochemistry 1990, 29, 955–960. [Google Scholar] [CrossRef]
- Schleep, S.; Kolodziej, H. Phenolic metabolites from Potentilla erecta. Phytochemistry (Life Sci. Adv.) 1992, 11, 87–92. [Google Scholar]
- Kolodziej, H. Procyanidins from medicinal birch: Bonding patterns and sequence of units in triflavanoids of mixed stereochemistry. Phytochemistry 1989, 28, 3487–3492. [Google Scholar] [CrossRef]
- Viviers, P.; Kolodziej, H.; Young, D.; Ferreira, D.; Roux, D. Synthesis of condensed tannins. Part 11. Intramolecular enantiomerism of the constituent units of tannins from the anacardiaceae: Stoicheiometric control in direct synthesis: Derivation of 1H-nuclear magnetic resonance parameters applicable to higher oligomers. J. Chem. Soc. Perkin Trans. 1 1983, 527–533. [Google Scholar] [CrossRef]
- Li, C.; Leverence, R.; Trombley, J.D.; Xu, S.; Yang, J.; Tian, Y.; Reed, J.D.; Hagerman, A.E. High molecular weight persimmon (Diospyros kaki L.) proanthocyanidin: A highly galloylated, A-linked tannin with an unusual flavonol terminal unit, myricetin. J. Agric. Food Chem. 2010, 58, 9033–9042. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, I.; Connaris, H.; Taylor, M.; Zaitsev, V.; Wilson, J.C.; Kiefel, M.J.; Itzstein, M.; Taylor, G. Sialic acid recognition by Vibrio cholerae neuraminidase. J. Biol. Chem. 2004, 279, 40819–40826. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.; Darowski, N.; Fuchs, M.R.; Förster, R.; Hellmig, M.; Paithankar, K.S.; Pühringer, S.; Steffien, M.; Zocher, G.; Weiss, M.S. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Radiat. 2012, 19, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Sparta, K.M.; Krug, M.; Heinemann, U.; Mueller, U.; Weiss, M.S. XDSAPP2.0. J. Appl. Cryst. 2016, 49, 1085–1092. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Storoni, L.C.; Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. Sect. D 2005, 61, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Schüttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Krissinell, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. TEXshade: Shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| Test Substance | IC50 (Viral) µg/mL µM | IC50 (Bacterial) µg/mL µM | ||
|---|---|---|---|---|
| Positive control | ||||
| oseltamivir carboxylate | 2.9 ± 0.2 (1) | 0.01 ± 0.001 | 41 ± 1 | 144 ± 1 |
| zanamivir | 3.7 ± 0.4 (1) | 0.01 ± 0.001 | 17 ± 1 | 52 ± 2 |
| Flavan-3-ols | ||||
| catechin | 312 ± 21 | 1076 ± 75 | 595 ± 25 | 2050 ± 87 |
| gallocatechin | 547 ± 23 | 1787 ± 74 | 603 ± 61 | 1969 ± 199 |
| catechin-3-O-gallate | 862 ± 2 | 1949 ± 4 | 24 ± 2 | 55 ± 4 |
| gallocatechin-3-O-gallate | 181 ± 3 | 396 ± 7 | 11 ± 1 | 25 ± 2 |
| epicatechin | 305 ± 19 | 1053 ± 64 | 670 ± 29 | 2186 ± 99 |
| epigallocatechin | 532 ± 41 | 1739 ± 135 | 598 ± 57 | 1955 ± 185 |
| epicatechin-3-O-gallate | 845 ± 24 | 1910 ± 55 | 93 ± 8 | 211 ± 19 |
| epigallocatechin-3-O-gallate | 717 ± 63 | 1565 ± 137 | 29 ± 1 | 64 ± 3 |
| Test Substance | IC50 (Viral) µg/mL µM | IC50 (Bacterial) µg/mL µM | ||
|---|---|---|---|---|
| Positive control | ||||
| oseltamivir acid | 2.9 ± 0.2 (1) | 0.01 ± 0.001 | 41 ± 1 | 144 ± 1 |
| zanamivir | 3.7 ± 0.4 (1) | 0.01 ± 0.001 | 17 ± 1 | 52 ± 2 |
| Ellagitannins | ||||
| dehydroellagitannin members | ||||
| geraniin | - | - | 128 ± 2 | 135 ± 2 |
| granatin A | - | - | 124 ± 2 | 158 ± 4 |
| carpinusin | - | - | 131 ± 5 | 138 ± 5 |
| terchebin | 97 ± 2 | 101 ± 3 | 29 ± 2 | 31 ± 2 |
| C-glycosidic members | ||||
| casuariin | - | - | 185 ± 4 | 236 ± 5 |
| vescalagin | - | - | 125 ± 11 | 73 ± 11 |
| paeonianin C | 587 ± 24 | 344 ± 14 | 25 ± 2 | 15 ± 1 |
| Tested Substance | Constituent Flavanyl Units | IC50 (Viral) µg/mL | IC50 (Bacterial) µg/mL |
|---|---|---|---|
| Positive control | |||
| oseltamivir acid | 2.9 ± 0.2 (1) | 41 ± 1 | |
| zanamivir | 3.7 ± 0.4 (1) | 17 ± 1 | |
| Tannin fractions | |||
| Diospyros kaki | galloylated flavan-3-ols | 20 ± 1 | 0.5 ± 0.04 |
| EPs® 7630 (2) | (epi)gallocatechin/(epi)catechin | 61 ± 2 | 1.7 ± 0.1 |
| Nelia meyeri | epicatechin | 29 ± 1 | 3.2 ± 0.1 |
| Salix ssp. | catechin | 32 ± 3 | 4.4 ± 0.2 |
| Potentilla erecta | epicatechin/catechin | - | 9.2 ± 1 |
| Betula spp. | epicatechin/catechin | - | 13 ± 1 |
| Rhus leptodictya | fisetinidol | - | 25 ± 1 |
| Compound | Concentration–Effect Parameters | CI Values at | CIwt | Combined Effect | ||||
|---|---|---|---|---|---|---|---|---|
| Dm | m | IC50 | IC75 | IC90 | IC95 | |||
| zanamivir | 17.0 ± 1 | 0.9 | - | - | - | - | - | |
| EPs® 7630 | 1.7 ± 0.1 | 1.4 | - | - | - | - | - | |
| Ratio of Zanamivir and EPs® 7630 | ||||||||
| 5:1 | 7.8 ± 0.6 | 1.1 | 1.1 | 0.7 | 0.5 | 0.4 | 0.5 | synergistic |
| 10:1 | 9.1 ± 0.4 | 1.7 | 0.9 | 0.7 | 0.6 | 0.5 | 0.6 | synergistic |
| 1:1 | 7.8 ± 0.6 | 1.7 | 2.3 | 1.8 | 1.5 | 1.3 | 1.6 | antagonistic |
| 1:5 | 1.4 ± 0.1 | 1.2 | 0.7 | 0.8 | 1.0 | 1.1 | 1.0 | additive |
| 1:10 | 1.4 ± 0.2 | 1.0 | 0.7 | 0.8 | 0.8 | 0.9 | 0.8 | moderate synergistic |
| Oseltamivir Carboxylate | Zanamivir | |
|---|---|---|
| Data Collection | ||
| Space group | P212121 | C121 |
| Cell dimension | ||
| a, b, c (Å) | 71.76, 77.86, 163.48 | 190.55, 50.34, 86.09 |
| α, β, γ (˚) | 90, 90, 90 | 90, 107.24, 90 |
| Resolution (Å) | 44.33–1.87 (1.94–1.87) * | 46.00–1.75 (1.81–1.75) |
| Rmerge (%) | 10.1 (49.6) | 4.4 (47.8) |
| <I/σ(I)> | 10.2 (2.4) | 14.7 (1.8) |
| Completeness (%) | 99.2 (97.9) | 97.8 (96.8) |
| Multiplicity | 3.7 (3.5) | 2.3 (2.2) |
| Refinement | ||
| Resolution (Å) | 44.33–1.87 | 46.00–1.75 |
| No. reflections | 282260 | 176727 |
| Rwork/Rfree (%) | 16.31/20.44 | 16.69/20.29 |
| No. atoms | 6632 | 6569 |
| Protein | 5834 | 5852 |
| Ligand/ion | 42 | 57 |
| Water | 756 | 660 |
| Average B-factor (Å2) | ||
| Overall | 24.3 | 32.0 |
| Protein | 23.4 | 31.3 |
| Ligand/ion | 22.7 | 36.0 |
| Water | 31.9 | 38.0 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.007 |
| Bond angles (˚) | 1.18 | 1.05 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quosdorf, S.; Schuetz, A.; Kolodziej, H. Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay. Molecules 2017, 22, 1989. https://doi.org/10.3390/molecules22111989
Quosdorf S, Schuetz A, Kolodziej H. Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay. Molecules. 2017; 22(11):1989. https://doi.org/10.3390/molecules22111989
Chicago/Turabian StyleQuosdorf, Stefanie, Anja Schuetz, and Herbert Kolodziej. 2017. "Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay" Molecules 22, no. 11: 1989. https://doi.org/10.3390/molecules22111989