PSN-PC: A Novel Antimicrobial and Anti-Biofilm Peptide from the Skin Secretion of Phyllomedusa-camba with Cytotoxicity on Human Lung Cancer Cell
Abstract
:1. Introduction
2. Results
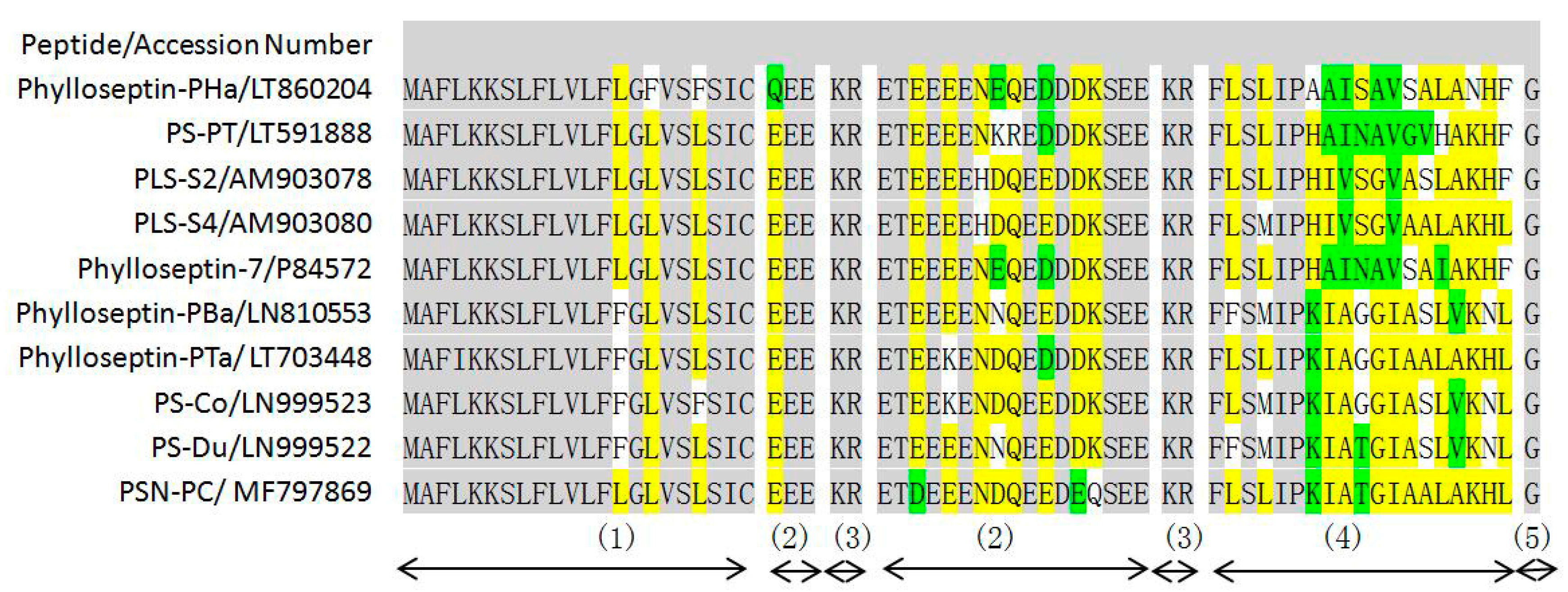
2.1. Molecular Cloning of a Novel AMP Precursor-Encoding cDNA and Bioinformatic Analyses
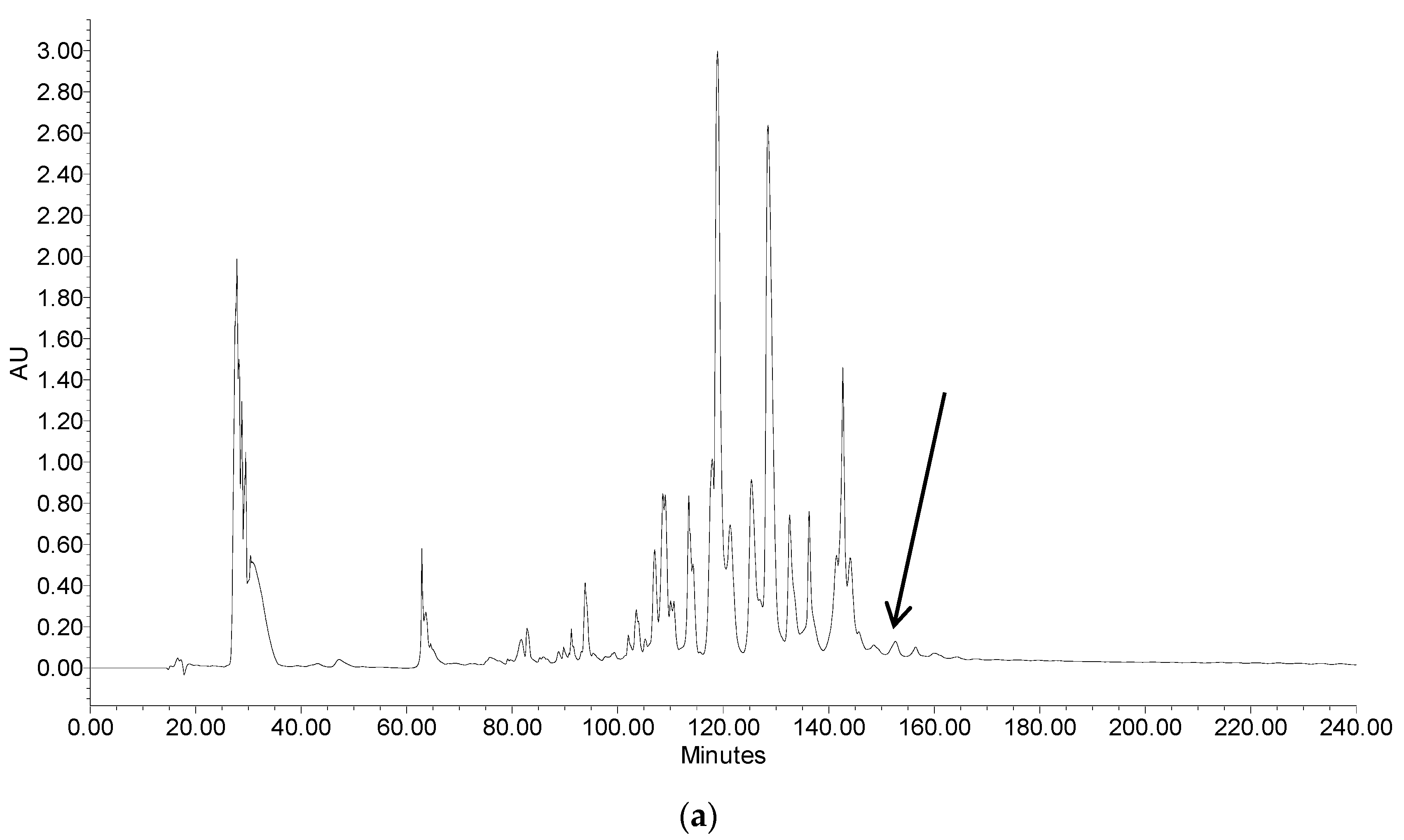
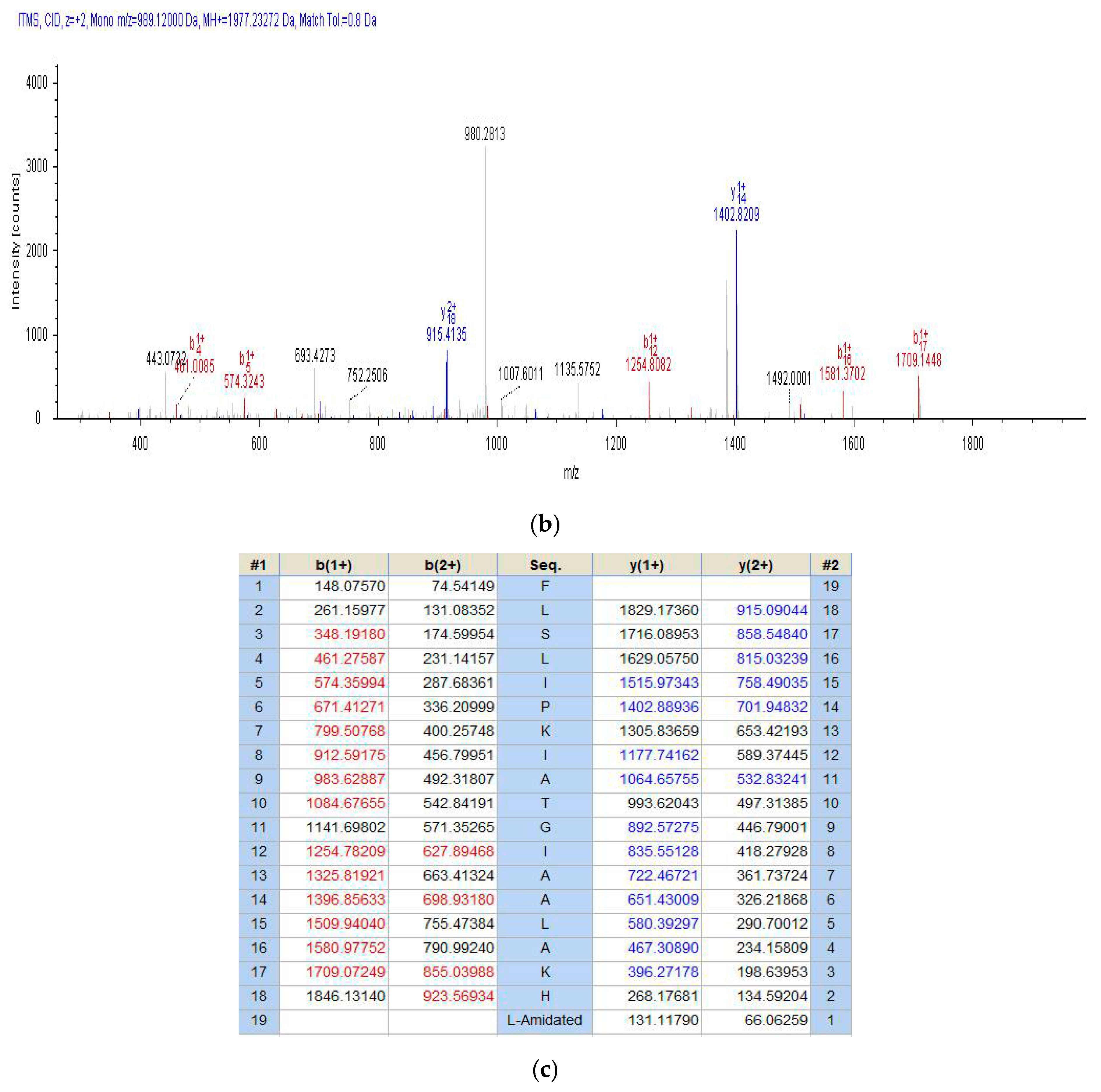
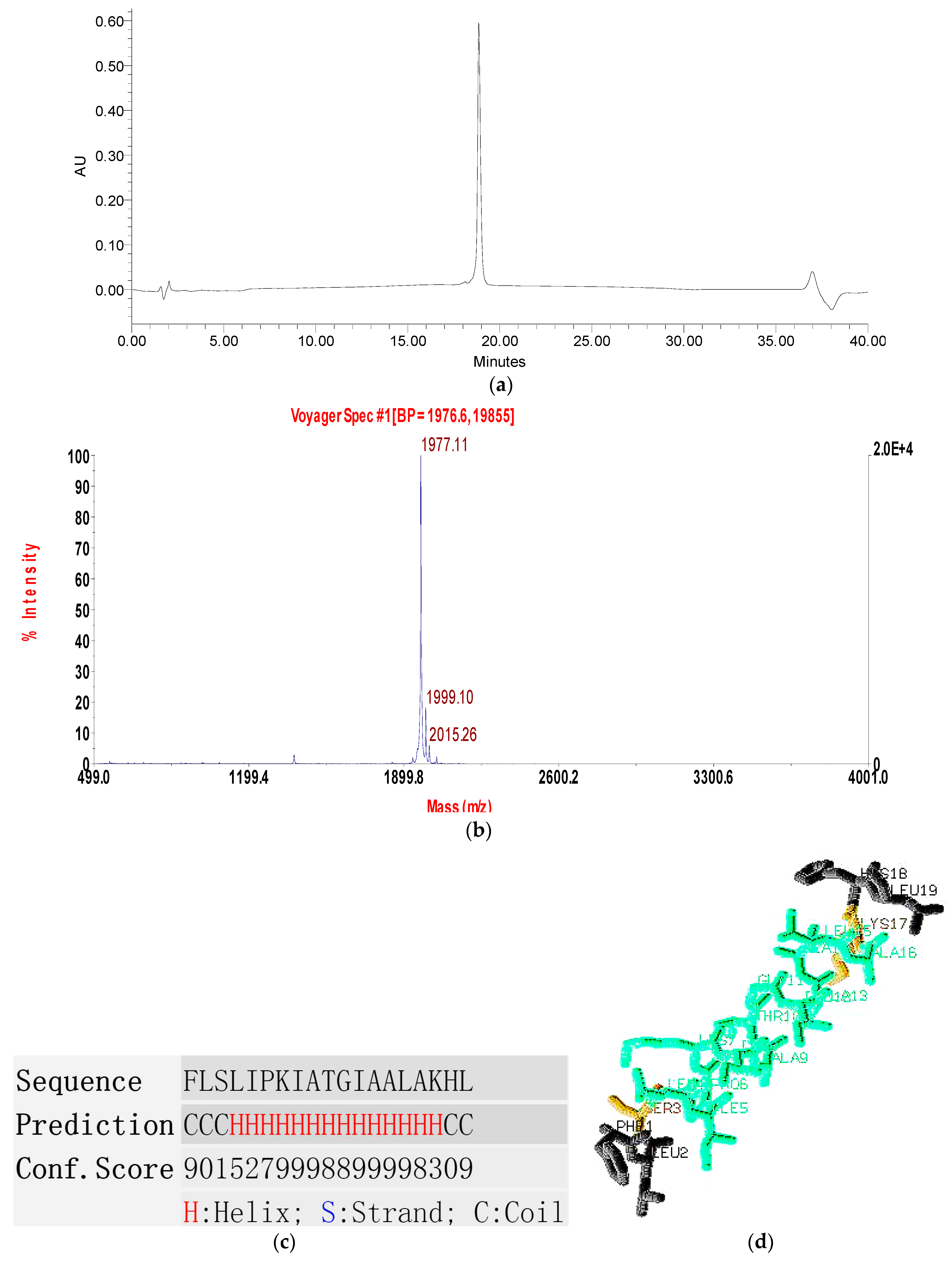
2.2. Fractionation of Skin Secretion, Identification and Structural Characterisation of PSN-PC
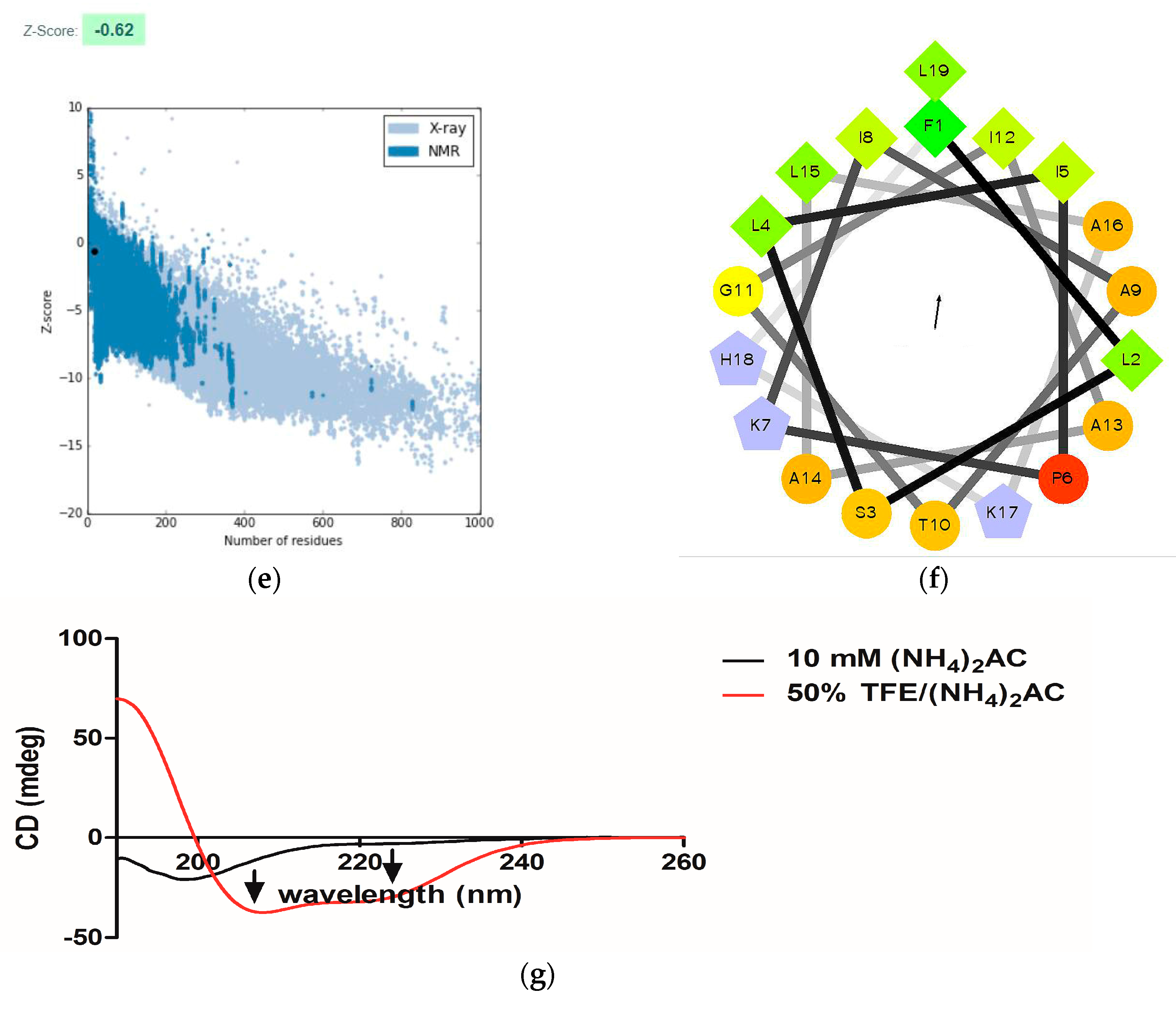
2.3. Conformational Study
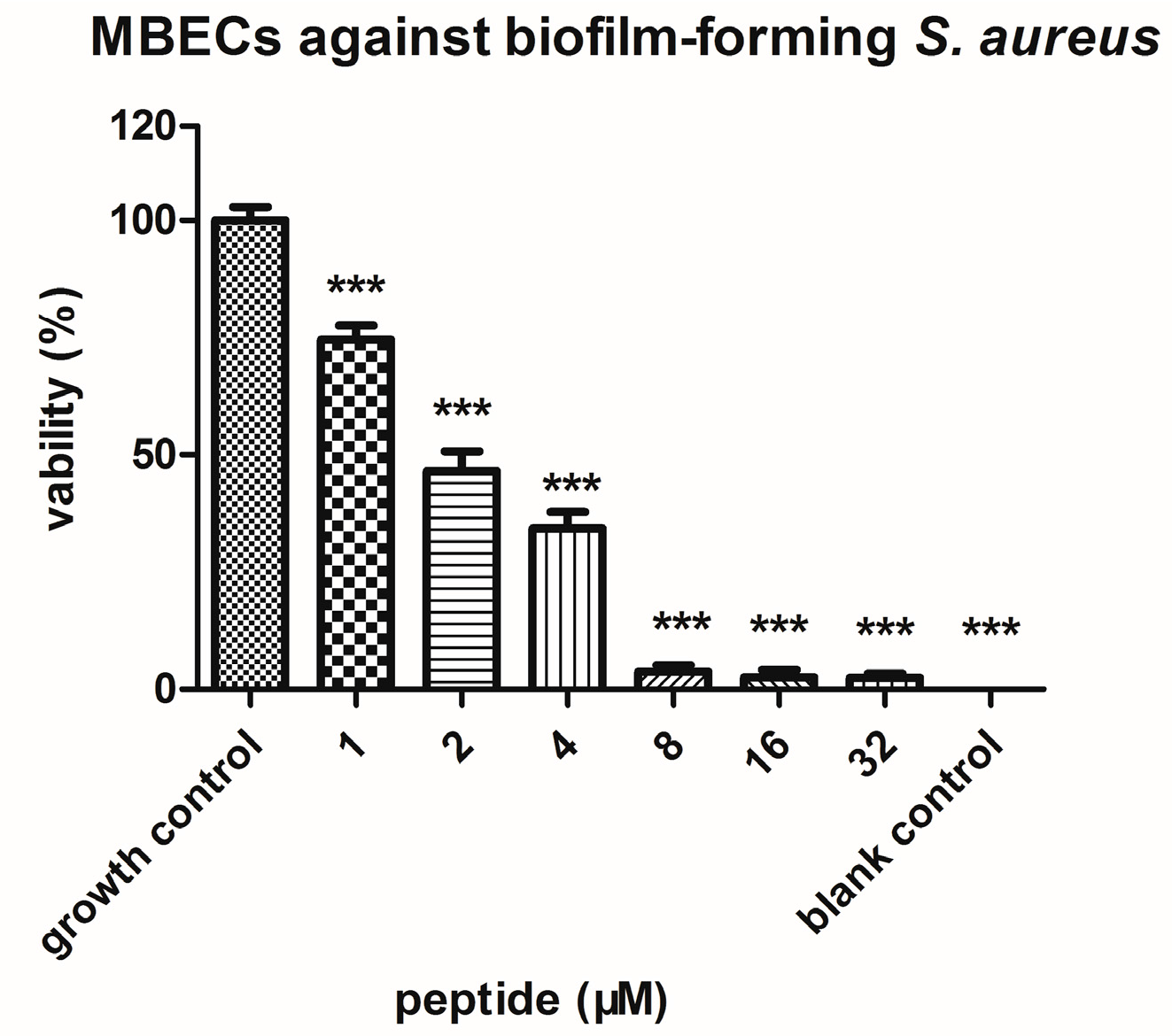
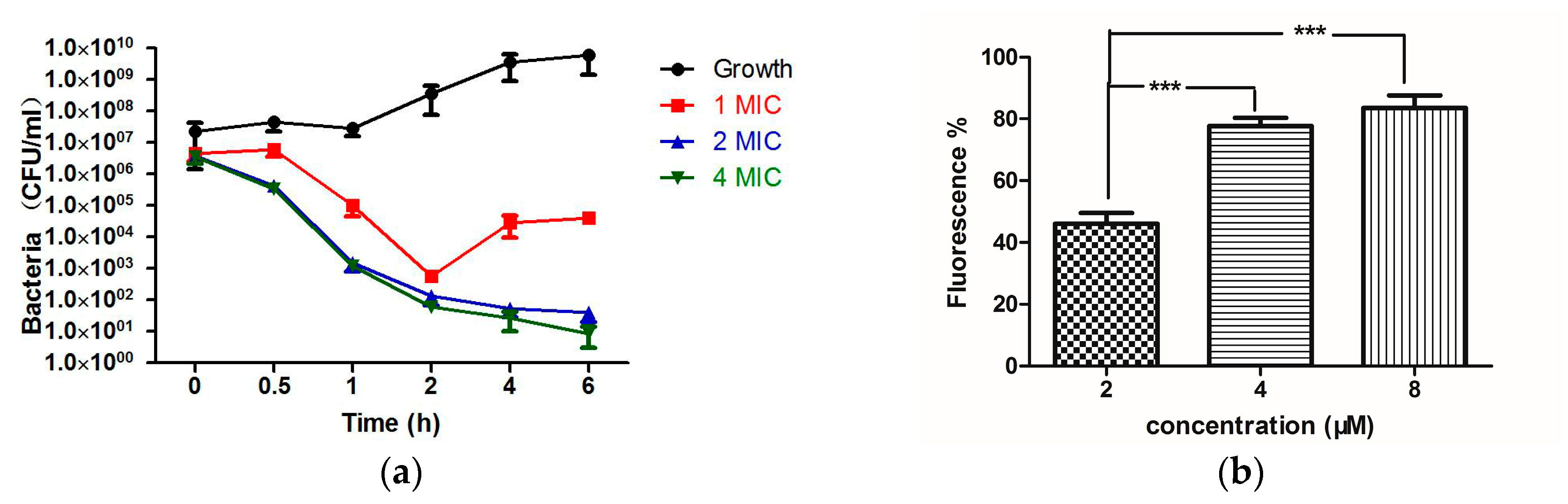
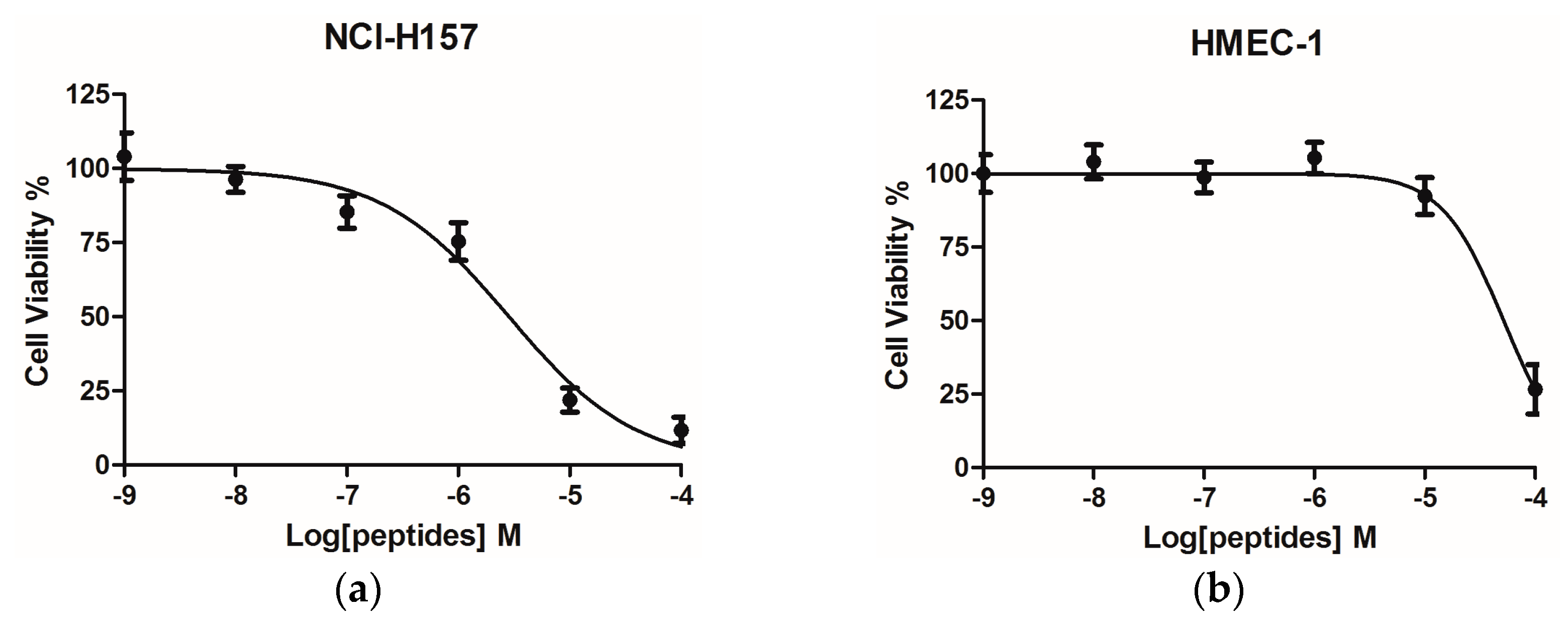
2.4. Bioactivity Assays of PSN-PC
3. Discussion
4. Materials and Methods
4.1. Acquisition of Skin Secretion
4.2. “Shotgun” Cloning of a Phyllomedusa camba Skin Secretion-Derived cDNA Library
4.3. Identification and Structural Analysis of PSN-PC
4.4. Solid-Phase Peptide Synthesis
4.5. Circular Dichroism (CD) Spectroscopy
4.6. Antimicrobial Activities
4.7. Anti-Biofilm Assays with S. aureus
4.8. Time-Killing Assay with S. aureus
4.9. Haemolytic Assay
4.10. Bacterial Cell Membrane Permeability Assay of PSN-PC Using S. aureus
4.11. Cells Lines and Cell Culture
4.12. Assessment of Cancer Cell Anti-Proliferative Activity Using the MTT Cell Viability Assay
4.13. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AMPs | antimicrobial peptide |
| CD | circular dichroism |
| DMEM | Dulbecco’s modified eagle’s medium |
| FBS | fetal bovine serum |
| MALDI-TOF MS | matrix-assisted laser desorption/ionization time of flight mass spectrometry |
| MBC | minimum bactericidal concentration |
| MBEC | minimal biofilm eradication concentration |
| MHA | Mueller-Hinton agar |
| MHB | Mueller-Hinton broth |
| MIC | minimum inhibitory concentration |
| MS/MS | tandem mass spectrometry |
| NUP | nested universal primer |
| PBS | phosphate-buffered saline |
| RP-HPLC | reverse-phase high performance liquid chromatography |
| TFA | trifluoroacetic acid |
| TFE | trifluoroethanol |
| TIPS | triisopropylsilane |
| TSB | tryptic soy broth |
| TTC | 2,3,5-triphenyl tetrazolium chloride |
References
- Kückelhaus, S.A.; Leite, J.R.S.; Muniz-Junqueira, M.I.; Sampaio, R.N.; Bloch, C.; Tosta, C.E. Antiplasmodial and antileishmanial activities of phylloseptin-1, an antimicrobial peptide from the skin secretion of Phyllomedusa azurea (Amphibia). Exp. Parasitol. 2009, 123, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Lukic, M.L.; Flatt, P.R. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides 2014, 57, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Zairi, A.; Belaïd, A.; Gahbiche, A.; Hani, K. Spermicidal activity of dermaseptins. Contraception 2005, 72, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Ageitos, J.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T. Antimicrobial peptides (amps): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Grassi, L.; Maisetta, G.; Maccari, G.; Esin, S.; Batoni, G. Analogs of the frog-skin antimicrobial peptide temporin 1tb exhibit a wider spectrum of activity and a stronger antibiofilm potential as compared to the parental peptide. Front. Chem. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Batista, C.; da Silva, L.R.; Sebben, A.; Scaloni, A.; Ferrara, L.; Paiva, G.; Olamendi–Portugal, T.; Possani, L.; Bloch, C. Antimicrobial peptides from the brazilian frog Phyllomedusa distincta. Peptides 1999, 20, 679–686. [Google Scholar] [CrossRef]
- Leite, J.R.S.; Silva, L.P.; Rodrigues, M.I.S.; Prates, M.V.; Brand, G.D.; Lacava, B.M.; Azevedo, R.B.; Bocca, A.L.; Albuquerque, S.; Bloch, C. Phylloseptins: A novel class of anti-bacterial and anti-protozoan peptides from the Phyllomedusa genus. Peptides 2005, 26, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, Y.H.; Power, G.J.; Flatt, P.R.; Woodhams, D.C.; Rollins-Smith, L.A.; Conlon, J.M. A peptide of the phylloseptin family from the skin of the frog Hylomantis lemur (Phyllomedusinae) with potent in vitro and in vivo insulin-releasing activity. Peptides 2008, 29, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Ma, C.; Zhou, M.; Xi, X.; Li, L.; Wu, D.; Wang, L.; Lin, C.; Lopez, J.C.; Chen, T. Phylloseptin-PBa—A novel broad-spectrum antimicrobial peptide from the skin secretion of the peruvian purple-sided leaf frog (Phyllomedusa Baltea) which exhibits cancer cell cytotoxicity. Toxins 2015, 7, 5182–5193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conceição, K.; Konno, K.; Richardson, M.; Antoniazzi, M.M.; Jared, C.; Daffre, S.; Camargo, A.C.M.; Pimenta, D.C. Isolation and biochemical characterization of peptides presenting antimicrobial activity from the skin of Phyllomedusa hypochondrialis. Peptides 2006, 27, 3092–3099. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Li, L.; Wu, D.; Gao, Y.; Xi, X.; Zhou, M.; Wang, L.; Chen, T.; Shaw, C. Discovery of novel bacterial cell-penetrating phylloseptins in defensive skin secretions of the south american hylid frogs, Phyllomedusa duellmani and Phyllomedusa coelestis. Toxins 2016, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, D.; Xi, X.; Wu, Y.; Ma, C.; Zhou, M.; Wang, L.; Yang, M.; Chen, T.; Shaw, C. Identification and characterisation of the antimicrobial peptide, phylloseptin-pt, from the skin secretion of Phyllomedusa tarsius, and comparison of activity with designed, cationicity-enhanced analogues and diastereomers. Molecues 2016, 21, 1667. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, Q.; Li, L.; Xi, X.; Wu, D.; Zhou, M.; Chen, T.; Shaw, C.; Wang, L. Discovery of phylloseptins that defense against gram-positive bacteria and inhibit the proliferation of the non-small cell lung cancer cell line, from the skin secretions of Phyllomedusa frogs. Molecues 2017, 22, 1428. [Google Scholar] [CrossRef] [PubMed]
- Raja, Z.; Andre, S.; Piesse, C.; Sereno, D.; Nicolas, P.; Foulon, T.; Oury, B.; Ladram, A. Structure, antimicrobial activities and mode of interaction with membranes of bovel phylloseptins from the painted-belly leaf frog, Phyllomedusa sauvagii. PLoS ONE 2013, 8, e70782. [Google Scholar] [CrossRef]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 2003, 43, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Leonardo de Azevedo Calderon, L.; Alexandre de Almeida, E.S.; Ciancaglini, P.; Stábeli, R.G. Antimicrobial peptides from Phyllomedusa frogs: From biomolecular diversity to potential nanotechnologic medical applications. Amino Acids 2011, 40, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Amiche, M.; Ladram, A.; Nicolas, P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides 2008, 29, 2074–2082. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.F.; Soto, A.; Knoop, F.C.; Conlon, J.M. Antimicrobial peptides isolated from skin secretions of the diploid frog, xenopus tropicalis (pipidae). Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1550, 81–89. [Google Scholar] [CrossRef]
- Lee, J.K.; Gopal, R.; Park, S.-C.; Ko, H.S.; Kim, Y.; Hahm, K.-S.; Park, Y. A proline-hinge alters the characteristics of the amphipathic α-helical amps. PLoS ONE 2013, 8, e67597. [Google Scholar] [CrossRef] [PubMed]
- Khara, J.S.; Obuobi, S.; Wang, Y.; Hamilton, M.S.; Robertson, B.D.; Newton, S.M.; Yang, Y.Y.; Langford, P.R.; Ee, P.L.R. Disruption of drug-resistant biofilms using de novo designed short α-helical antimicrobial peptides with idealized facial amphiphilicity. Acta Biomater. 2017, 57, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Henzler Wildman, K.A.; Lee, D.-K.; Ramamoorthy, A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, ll-37. Biochemistry 2003, 42, 6545–6558. [Google Scholar] [CrossRef] [PubMed]
- Hallock, K.J.; Lee, D.-K.; Ramamoorthy, A. Msi-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys. J. 2003, 84, 3052–3060. [Google Scholar] [CrossRef]
- McPhee, J.B.; Scott, M.G.; Hancock, R.E. Design of host defence peptides for antimicrobial and immunity enhancing activities. Comb. Chem. High. Throughput Screen. 2005, 8, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Park, S.C.; Park, H.K.; Shin, S.Y.; Kim, Y.; Hahm, K.S. Structure-activity relationship of hp (2–20) analog peptide: Enhanced antimicrobial activity by n-terminal random coil region deletion. Pept. Sci. 2007, 88, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kazemzadeh-Narbat, M.; Kindrachuk, J.; Duan, K.; Jenssen, H.; Hancock, R.E.; Wang, R. Antimicrobial peptides on calcium phosphate-coated titanium for the prevention of implant-associated infections. Biomaterials 2010, 31, 9519–9526. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.K.; Abouelhassan, Y.; Zuo, R.; Yousaf, H.; Ding, Y.; Huigens, R.W. Antimicrobial peptide-inspired nh125 analogues: Bacterial and fungal biofilm-eradicating agents and rapid killers of mrsa persisters. Org. Biomol. Chem. 2017, 15, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Schröder-Borm, H.; Bakalova, R.; Andrä, J. The nk-lysin derived peptide nk-2 preferentially kills cancer cells with increased surface levels of negatively charged phosphatidylserine. FEBS Lett. 2005, 579, 6128–6134. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.; Hamadat, S.; Le Saux, K.; Newton, C.; Mazouni, M.; Zargarian, L.; Miro-Padovani, M.; Zadigue, P.; Delbé, J.; Hamma-Kourbali, Y. Studies of the antitumor mechanism of action of dermaseptin b2, a multifunctional cationic antimicrobial peptide, reveal a partial implication of cell surface glycosaminoglycans. PLoS ONE 2017, 12, e0182926. [Google Scholar] [CrossRef] [PubMed]
- Bocchinfuso, G.; Palleschi, A.; Orioni, B.; Grande, G.; Formaggio, F.; Toniolo, C.; Park, Y.; Hahm, K.S.; Stella, L. Different mechanisms of action of antimicrobial peptides: Insights from fluorescence spectroscopy experiments and molecular dynamics simulations. J. Pept. Sci. 2009, 15, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.; Liu, L.-P.; Deber, C.M. Cationic hydrophobic peptides with antimicrobial activity. Antimicrob. Agents Chemother. 2002, 46, 3585–3590. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, M.; Gagliardo, R.; Walker, B.; Shaw, C. Elements of the granular gland peptidome and transcriptome persist in air-dried skin of the south american orange-legged leaf frog, Phyllomedusa hypocondrialis. Peptides 2006, 27, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shaw, C. Identification and molecular cloning of novel trypsin inhibitor analogs from the dermal venom of the oriental fire-bellied toad (bombina orientalis) and the european yellow-bellied toad (bombina variegata). Peptides 2003, 24, 873–880. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-tasser: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The i-tasser suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinf. 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. Prosa-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (mic) of antimicrobial substances. Nat. Protoc. 2008, 3, 163. [Google Scholar] [CrossRef] [PubMed]
- Sabaeifard, P.; Abdi-Ali, A.; Soudi, M.R.; Dinarvand, R. Optimization of tetrazolium salt assay for pseudomonas aeruginosa biofilm using microtiter plate method. J. Microbiol. Methods 2014, 105, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.L.; Poot, M.; Yue, S.T.; Millard, P.J. Bacterial viability and antibiotic susceptibility testing with sytox green nucleic acid stain. Appl. Environ. Microbiol. 1997, 63, 2421–2431. [Google Scholar] [PubMed]
- Pan, Y.; Wan, J.; Roginski, H.; Lee, A.; Shiell, B.; Michalski, W.; Coventry, M. Comparison of the effects of acylation and amidation on the antimicrobial and antiviral properties of lactoferrin. Lett. Appl. Microbiol. 2007, 44, 229–234. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds PSN-PC are not available from the authors. |


| Strains | S. aureus | MRSA | C. albicans | E. coli | P. aeruginosa |
|---|---|---|---|---|---|
| MIC (µM) | 2 | 2 | 2 | 8 | 32 |
| MBC (µM) | 4 | 4 | 2 | 8 | 64 |
| Peptide Name | MIC (mg.L−1/µM) | <H> | <µH> | Net Charge | ||
|---|---|---|---|---|---|---|
| S. aureus | E. coli | C. albicans | ||||
| Phylloseptin-PHa | 64/33 | >512/>264 | 256/131.9 | 0.799 | 0.457 | 0 |
| PS-PT | 55/26.4 | 55/26.4 | 55/26.4 | 0.686 | 0.438 | + 2 |
| PLS-S2 | 12.7/6.3 | 50.9/25 | ND | 0.801 | 0.548 | + 1 |
| PLS-S4 | 12.5/6.3 | 50.1/25 | ND | 0.789 | 0.519 | + 1 |
| Phylloseptin-7 | 12/6 | 12/6 | ND | 0.745 | 0.513 | + 1 |
| Phylloseptin-PBa | 8/4.2 | 128/67.6 | 8/4.2 | 0.711 | 0.637 | + 2 |
| Phylloseptin-PTa | 8/4.1 | 32/16.6 | 4/2.1 | 0.740 | 0.577 | + 2 |
| PS-Co | 8/4.1 | 128/64.9 | 16/8.1 | 0.706 | 0.636 | + 2 |
| PS-Du | 8/3.90 | 128/62.5 | 16/7.8 | 0.725 | 0.624 | + 2 |
| PSN-PC | 4/2 | 16/8 | 4/2 | 0.754 | 0.563 | + 2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Pan, J.; Wu, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Chen, T. PSN-PC: A Novel Antimicrobial and Anti-Biofilm Peptide from the Skin Secretion of Phyllomedusa-camba with Cytotoxicity on Human Lung Cancer Cell. Molecules 2017, 22, 1896. https://doi.org/10.3390/molecules22111896
Wu X, Pan J, Wu Y, Xi X, Ma C, Wang L, Zhou M, Chen T. PSN-PC: A Novel Antimicrobial and Anti-Biofilm Peptide from the Skin Secretion of Phyllomedusa-camba with Cytotoxicity on Human Lung Cancer Cell. Molecules. 2017; 22(11):1896. https://doi.org/10.3390/molecules22111896
Chicago/Turabian StyleWu, Xianhui, Jinhuo Pan, Yue Wu, Xinping Xi, Chengbang Ma, Lei Wang, Mei Zhou, and Tianbao Chen. 2017. "PSN-PC: A Novel Antimicrobial and Anti-Biofilm Peptide from the Skin Secretion of Phyllomedusa-camba with Cytotoxicity on Human Lung Cancer Cell" Molecules 22, no. 11: 1896. https://doi.org/10.3390/molecules22111896




